Conformational flexibility of N-glycans in solution studied by REMD simulations
- PMID: 28510079
- PMCID: PMC5418406
- DOI: 10.1007/s12551-012-0090-y
Conformational flexibility of N-glycans in solution studied by REMD simulations
Abstract
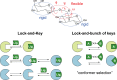
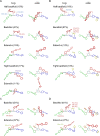
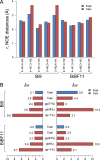
Protein-glycan recognition regulates a wide range of biological and pathogenic processes. Conformational diversity of glycans in solution is apparently incompatible with specific binding to their receptor proteins. One possibility is that among the different conformational states of a glycan, only one conformer is utilized for specific binding to a protein. However, the labile nature of glycans makes characterizing their conformational states a challenging issue. All-atom molecular dynamics (MD) simulations provide the atomic details of glycan structures in solution, but fairly extensive sampling is required for simulating the transitions between rotameric states. This difficulty limits application of conventional MD simulations to small fragments like di- and tri-saccharides. Replica-exchange molecular dynamics (REMD) simulation, with extensive sampling of structures in solution, provides a valuable way to identify a family of glycan conformers. This article reviews recent REMD simulations of glycans carried out by us or other research groups and provides new insights into the conformational equilibria of N-glycans and their alteration by chemical modification. We also emphasize the importance of statistical averaging over the multiple conformers of glycans for comparing simulation results with experimental observables. The results support the concept of "conformer selection" in protein-glycan recognition.
Keywords: Conformational flexibility; Conformer selection; Molecular dynamics simulations; N-glycan; N-glycan modifications; Protein–glycan interactions; Replica-exchange molecular dynamics simulations.
Figures


Similar articles
-
Effect of bisecting GlcNAc and core fucosylation on conformational properties of biantennary complex-type N-glycans in solution.J Phys Chem B. 2012 Jul 26;116(29):8504-12. doi: 10.1021/jp212550z. Epub 2012 May 2. J Phys Chem B. 2012. PMID: 22530754
-
Structural diversity and changes in conformational equilibria of biantennary complex-type N-glycans in water revealed by replica-exchange molecular dynamics simulation.Biophys J. 2011 Nov 16;101(10):L44-6. doi: 10.1016/j.bpj.2011.10.019. Epub 2011 Nov 15. Biophys J. 2011. PMID: 22098756 Free PMC article.
-
Replica-Exchange Methods for Biomolecular Simulations.Methods Mol Biol. 2019;2022:155-177. doi: 10.1007/978-1-4939-9608-7_7. Methods Mol Biol. 2019. PMID: 31396903
-
Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms.Biochim Biophys Acta. 2016 Jul;1858(7 Pt B):1635-51. doi: 10.1016/j.bbamem.2015.12.032. Epub 2016 Jan 5. Biochim Biophys Acta. 2016. PMID: 26766517 Free PMC article. Review.
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
Cited by
-
Structural and Functional Insights into GluK3-kainate Receptor Desensitization and Recovery.Sci Rep. 2019 Jul 16;9(1):10254. doi: 10.1038/s41598-019-46770-z. Sci Rep. 2019. PMID: 31311973 Free PMC article.
-
Conformational Heterogeneity of the HIV Envelope Glycan Shield.Sci Rep. 2017 Jun 30;7(1):4435. doi: 10.1038/s41598-017-04532-9. Sci Rep. 2017. PMID: 28667249 Free PMC article.
-
Multiscale Simulations Examining Glycan Shield Effects on Drug Binding to Influenza Neuraminidase.Biophys J. 2020 Dec 1;119(11):2275-2289. doi: 10.1016/j.bpj.2020.10.024. Epub 2020 Oct 31. Biophys J. 2020. PMID: 33130120 Free PMC article.
-
Three-Dimensional Structures of Carbohydrates and Where to Find Them.Int J Mol Sci. 2020 Oct 18;21(20):7702. doi: 10.3390/ijms21207702. Int J Mol Sci. 2020. PMID: 33081008 Free PMC article. Review.
-
Conformational sampling of oligosaccharides using Hamiltonian replica exchange with two-dimensional dihedral biasing potentials and the weighted histogram analysis method (WHAM).J Chem Theory Comput. 2015 Feb 10;11(2):788-99. doi: 10.1021/ct500993h. J Chem Theory Comput. 2015. PMID: 25705140 Free PMC article.
References
-
- André S, Unverzagt C, Kojima S, Frank M, Seifert J, Fink C, Kayser K, von der Lieth CW, Gabius HJ. Determination of modulation of ligand properties of synthetic complex-type biantennary N-glycans by introduction of bisecting GlcNAc in silico, in vitro and in vivo. Eur J Biochem. 2004;271(1):118–134. doi: 10.1046/j.1432-1033.2003.03910.x. - DOI - PubMed
-
- Beckham GT, Bomble YJ, Matthews JF, Taylor CB, Resch MG, Yarbrough JM, Decker SR, Bu LT, Zhao XC, McCabe C, Wohlert J, Bergenstråhle M, Brady JW, Adney WS, Himmel ME, Crowley MF. The O-glycosylated linker from the trichoderma reesei family 7 cellulase is a flexible, disordered protein. Biophys J. 2010;99(11):3773–3781. doi: 10.1016/j.bpj.2010.10.032. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

