DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression
- PMID: 28510216
- PMCID: PMC5418507
- DOI: 10.1007/s12551-016-0238-2
DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression
Abstract
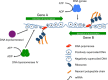
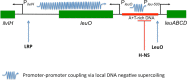
Although it has become routine to consider DNA in terms of its role as a carrier of genetic information, it is also an important contributor to the control of gene expression. This regulatory principle arises from its structural properties. DNA is maintained in an underwound state in most bacterial cells and this has important implications both for DNA storage in the nucleoid and for the expression of genetic information. Underwinding of the DNA through reduction in its linking number potentially imparts energy to the duplex that is available to drive DNA transactions, such as transcription, replication and recombination. The topological state of DNA also influences its affinity for some DNA binding proteins, especially in DNA sequences that have a high A + T base content. The underwinding of DNA by the ATP-dependent topoisomerase DNA gyrase creates a continuum between metabolic flux, DNA topology and gene expression that underpins the global response of the genome to changes in the intracellular and external environments. These connections describe a fundamental and generalised mechanism affecting global gene expression that underlies the specific control of transcription operating through conventional transcription factors. This mechanism also provides a basal level of control for genes acquired by horizontal DNA transfer, assisting microbial evolution, including the evolution of pathogenic bacteria.
Keywords: DNA supercoiling; DNA topoisomerases; Gene regulation; Transcription.
Conflict of interest statement
Conflict of interest
Charles J. Dorman declares that he has no conflict of interest.
Matthew J. Dorman declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

