Network modelling of gene regulation
- PMID: 28510232
- PMCID: PMC5418390
- DOI: 10.1007/s12551-010-0041-4
Network modelling of gene regulation
Abstract
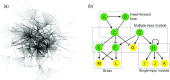
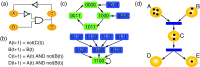
Gene regulatory network (GRN) modelling has gained increasing attention in the past decade. Many computational modelling techniques have been proposed to facilitate the inference and analysis of GRN. However, there is often confusion about the aim of GRN modelling, and how a gene network model can be fully utilised as a tool for systems biology. The aim of the present article is to provide an overview of this rapidly expanding subject. In particular, we review some fundamental concepts of systems biology and discuss the role of network modelling in understanding complex biological systems. Several commonly used network modelling paradigms are surveyed with emphasis on their practical use in systems biology research.
Keywords: Bioinformatics; Gene regulatory network; Systems biology.
Figures




References
-
- Aldana M. Boolean dynamics of networks with scale-free topology. Phys Nonlinear Phenom. 2003;185:45–66. doi: 10.1016/S0167-2789(03)00174-X. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

