Proteoforms in Peripheral Blood Mononuclear Cells as Novel Rejection Biomarkers in Liver Transplant Recipients
- PMID: 28510335
- PMCID: PMC5612406
- DOI: 10.1111/ajt.14359
Proteoforms in Peripheral Blood Mononuclear Cells as Novel Rejection Biomarkers in Liver Transplant Recipients
Abstract
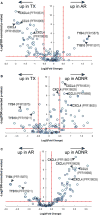
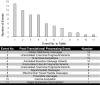
Biomarker profiles of acute rejection in liver transplant recipients could enhance the diagnosis and management of recipients. Our aim was to identify diagnostic proteoform signatures of acute rejection in circulating immune cells, using an emergent "top-down" proteomics methodology. We prepared differentially processed and cryopreserved cell lysates from 26 nonviral liver transplant recipients by molecular weight-based fractionation and analyzed them by mass spectrometry of whole proteins in three steps: (i) Nanocapillary liquid chromatography coupled with high-resolution tandem mass spectrometry; (ii) database searching to identify and characterize intact proteoforms; (iii) data processing through a hierarchical linear model matching the study design to quantify proteoform fold changes in patients with rejection versus normal liver function versus acute dysfunction without rejection. Differentially expressed proteoforms were seen in patients with rejection versus normal and nonspecific controls, most evidently in the cell preparations stored in traditional serum-rich media. Mapping analysis of these proteins back to genes through gene ontology and pathway analysis tools revealed multiple signaling pathways, including inflammation mediated by cytokines and chemokines. Larger studies are needed to validate these novel rejection signatures and test their predictive value for use in clinical management.
Keywords: biomarker; clinical research/practice; immunobiology; liver allograft function/dysfunction; liver transplantation/hepatology; monitoring: immune; proteomics; rejection: acute; translational research/science.
© 2017 The American Society of Transplantation and the American Society of Transplant Surgeons.
Conflict of interest statement
The authors of this manuscript have conflicts of interest to disclose as described by the
Figures


References
-
- Charlton M, Seaberg E. Impact of immunosuppression and acute rejection on recurrence of hepatitis C: Results of the national institute of diabetes and digestive and kidney diseases liver transplantation database. Liver Transpl Surg. 1999;5(4 Suppl 1):S107–S114. - PubMed
-
- Shaked A, Chang BL, Barnes MR, et al. An ectopically expressed serum miRNA signature is prognostic, diagnostic, and biologically related to liver allograft rejection. Hepatology. 2017;65:269–280. - PubMed
-
- Moya-Quiles MR, Alvarez R, Miras M, et al. Impact of recipient HLA-C in liver transplant: A protective effect of HLA-Cw*07 on acute rejection. Hum Immunol. 2007;68:51–58. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

