Human mismatch repair system balances mutation rates between strands by removing more mismatches from the lagging strand
- PMID: 28512192
- PMCID: PMC5538550
- DOI: 10.1101/gr.219915.116
Human mismatch repair system balances mutation rates between strands by removing more mismatches from the lagging strand
Abstract
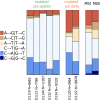
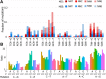
Mismatch repair (MMR) is one of the main systems maintaining fidelity of replication. Differences in correction of errors produced during replication of the leading and the lagging DNA strands were reported in yeast and in human cancers, but the causes of these differences remain unclear. Here, we analyze data on human cancers with somatic mutations in two of the major DNA polymerases, delta and epsilon, that replicate the genome. We show that these cancers demonstrate a substantial asymmetry of the mutations between the leading and the lagging strands. The direction of this asymmetry is the opposite between cancers with mutated polymerases delta and epsilon, consistent with the role of these polymerases in replication of the lagging and the leading strands in human cells, respectively. Moreover, the direction of strand asymmetry observed in cancers with mutated polymerase delta is similar to that observed in MMR-deficient cancers. Together, these data indicate that polymerase delta (possibly together with polymerase alpha) contributes more mismatches during replication than its leading-strand counterpart, polymerase epsilon; that most of these mismatches are repaired by the MMR system; and that MMR repairs about three times more mismatches produced in cells during lagging strand replication compared with the leading strand.
© 2017 Andrianova et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Chen C-L, Duquenne L, Audit B, Guilbaud G, Rappailles A, Baker A, Huvet M, d'Aubenton-Carafa Y, Hyrien O, Arneodo A, et al. 2011. Replication-associated mutational asymmetry in the human genome. Mol Biol Evol 28: 2327–2337. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
