Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes
- PMID: 28512350
- PMCID: PMC8127953
- DOI: 10.1038/nrm.2017.26
Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes
Abstract
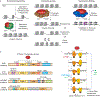
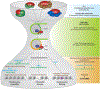
Cells utilize diverse ATP-dependent nucleosome-remodelling complexes to carry out histone sliding, ejection or the incorporation of histone variants, suggesting that different mechanisms of action are used by the various chromatin-remodelling complex subfamilies. However, all chromatin-remodelling complex subfamilies contain an ATPase-translocase 'motor' that translocates DNA from a common location within the nucleosome. In this Review, we discuss (and illustrate with animations) an alternative, unifying mechanism of chromatin remodelling, which is based on the regulation of DNA translocation. We propose the 'hourglass' model of remodeller function, in which each remodeller subfamily utilizes diverse specialized proteins and protein domains to assist in nucleosome targeting or to differentially detect nucleosome epitopes. These modules converge to regulate a common DNA translocation mechanism, to inform the conserved ATPase 'motor' on whether and how to apply DNA translocation, which together achieve the various outcomes of chromatin remodelling: nucleosome assembly, chromatin access and nucleosome editing.
Figures
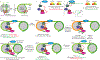
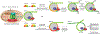
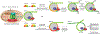

References
-
- Clapier CR & Cairns BR in Fundamentals of Chromatin (eds Workman JL & Abmayr SM) 69–146 (Springer, 2014).
-
- Wilson BG & Roberts CW SWI/SNF nucleosome remodellers and cancer. Nature reviews. Cancer 11, 481–492 (2011). - PubMed
-
- Clapier CR & Cairns BR The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273–304 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

