Protein S Negatively Regulates Neural Stem Cell Self-Renewal through Bmi-1 Signaling
- PMID: 28512399
- PMCID: PMC5411449
- DOI: 10.3389/fnmol.2017.00124
Protein S Negatively Regulates Neural Stem Cell Self-Renewal through Bmi-1 Signaling
Abstract
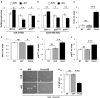
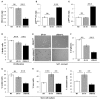
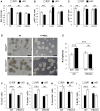
Revealing the molecular mechanisms underlying neural stem cell self-renewal is a major goal toward understanding adult brain homeostasis. The self-renewing potential of neural stem and progenitor cells (NSPCs) must be tightly regulated to maintain brain homeostasis. We recently reported the expression of Protein S (PROS1) in adult hippocampal NSPCs, and revealed its role in regulation of NSPC quiescence and neuronal differentiation. Here, we investigate the effect of PROS1 on NSPC self-renewal and show that genetic ablation of Pros1 in neural progenitors increased NSPC self-renewal by 50%. Mechanistically, we identified the upregulation of the polycomb complex protein Bmi-1 and repression of its downstream effectors p16Ink4a and p19Arf to promote NSPC self-renewal in Pros1-ablated cells. Rescuing Pros1 expression restores normal levels of Bmi-1 signaling, and reverts the proliferation and enhanced self-renewal phenotypes observed in Pros1-deleted cells. Our study identifies PROS1 as a novel negative regulator of NSPC self-renewal. We conclude PROS1 is instructive for NSPC differentiation by negatively regulating Bmi-1 signaling in adult and embryonic neural stem cells.
Keywords: Bmi-1; PROS1; Protein S; neural stem cells; neurogenesis; self-renewal.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

