Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life
- PMID: 28512451
- PMCID: PMC5411419
- DOI: 10.3389/fmicb.2017.00738
Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life
Abstract
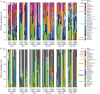
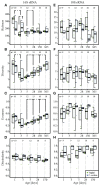
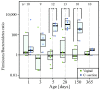
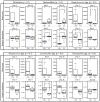
Perturbations to the colonization process of the human gastrointestinal tract have been suggested to result in adverse health effects later in life. Although much research has been performed on bacterial colonization and succession, much less is known about the other two domains of life, archaea, and eukaryotes. Here we describe colonization and succession by bacteria, archaea and microeukaryotes during the first year of life (samples collected around days 1, 3, 5, 28, 150, and 365) within the gastrointestinal tract of infants delivered either vaginally or by cesarean section and using a combination of quantitative real-time PCR as well as 16S and 18S rRNA gene amplicon sequencing. Sequences from organisms belonging to all three domains of life were detectable in all of the collected meconium samples. The microeukaryotic community composition fluctuated strongly over time and early diversification was delayed in infants receiving formula milk. Cesarean section-delivered (CSD) infants experienced a delay in colonization and succession, which was observed for all three domains of life. Shifts in prokaryotic succession in CSD infants compared to vaginally delivered (VD) infants were apparent as early as days 3 and 5, which were characterized by increased relative abundances of the genera Streptococcus and Staphylococcus, and a decrease in relative abundance for the genera Bifidobacterium and Bacteroides. Generally, a depletion in Bacteroidetes was detected as early as day 5 postpartum in CSD infants, causing a significantly increased Firmicutes/Bacteroidetes ratio between days 5 and 150 when compared to VD infants. Although the delivery mode appeared to have the strongest influence on differences between the infants, other factors such as a younger gestational age or maternal antibiotics intake likely contributed to the observed patterns as well. Our findings complement previous observations of a delay in colonization and succession of CSD infants, which affects not only bacteria but also archaea and microeukaryotes. This further highlights the need for resolving bacterial, archaeal, and microeukaryotic dynamics in future longitudinal studies of microbial colonization and succession within the neonatal gastrointestinal tract.
Keywords: amplicon sequencing; delivery mode; fungi; infant gut microbiome; microbial colonization; quantitative real-time PCR; succession.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

