Conversion of adult endothelium to immunocompetent haematopoietic stem cells
- PMID: 28514438
- PMCID: PMC5794215
- DOI: 10.1038/nature22326
Conversion of adult endothelium to immunocompetent haematopoietic stem cells
Abstract
Developmental pathways that orchestrate the fleeting transition of endothelial cells into haematopoietic stem cells remain undefined. Here we demonstrate a tractable approach for fully reprogramming adult mouse endothelial cells to haematopoietic stem cells (rEC-HSCs) through transient expression of the transcription-factor-encoding genes Fosb, Gfi1, Runx1, and Spi1 (collectively denoted hereafter as FGRS) and vascular-niche-derived angiocrine factors. The induction phase (days 0-8) of conversion is initiated by expression of FGRS in mature endothelial cells, which results in endogenous Runx1 expression. During the specification phase (days 8-20), RUNX1+ FGRS-transduced endothelial cells commit to a haematopoietic fate, yielding rEC-HSCs that no longer require FGRS expression. The vascular niche drives a robust self-renewal and expansion phase of rEC-HSCs (days 20-28). rEC-HSCs have a transcriptome and long-term self-renewal capacity similar to those of adult haematopoietic stem cells, and can be used for clonal engraftment and serial primary and secondary multi-lineage reconstitution, including antigen-dependent adaptive immune function. Inhibition of TGFβ and CXCR7 or activation of BMP and CXCR4 signalling enhanced generation of rEC-HSCs. Pluripotency-independent conversion of endothelial cells into autologous authentic engraftable haematopoietic stem cells could aid treatment of haematological disorders.
Figures
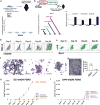





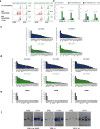


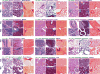



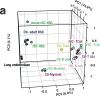


Comment in
-
Blood: Education for stem cells.Nature. 2017 May 25;545(7655):415-417. doi: 10.1038/nature22496. Epub 2017 May 17. Nature. 2017. PMID: 28514445 No abstract available.
References
-
- Wahlster L, Daley GQ. Progress towards generation of human haematopoietic stem cells. Nat. Cell Biol. 2016;18:1111–1117. - PubMed
-
- Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat. Biotechnol. 2013;31:416–418. - PubMed
-
- Tashiro K, et al. Promotion of hematopoietic differentiation from mouse induced pluripotent stem cells by transient HoxB4 transduction. Stem Cell Res. 2012;8:300–311. - PubMed
-
- Rafii S, et al. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. Blood. 2013;121:770–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL133021/HL/NHLBI NIH HHS/United States
- R01 CA204308/CA/NCI NIH HHS/United States
- HL128158/NH/NIH HHS/United States
- U01 HL099997/HL/NHLBI NIH HHS/United States
- R01 HL115128/HL/NHLBI NIH HHS/United States
- R01 HL091724/HL/NHLBI NIH HHS/United States
- HL099997/NH/NIH HHS/United States
- HL133021/NH/NIH HHS/United States
- U54 CA163167/CA/NCI NIH HHS/United States
- R01 HL128158/HL/NHLBI NIH HHS/United States
- R01 DK095039/DK/NIDDK NIH HHS/United States
- HL119872/NH/NIH HHS/United States
- R01 HL097797/HL/NHLBI NIH HHS/United States
- NIH-R01 HL091724/NH/NIH HHS/United States
- T32 HD060600/HD/NICHD NIH HHS/United States
- R01 HL119872/HL/NHLBI NIH HHS/United States
- HL115128/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

