Structure of the full-length glucagon class B G-protein-coupled receptor
- PMID: 28514451
- PMCID: PMC5492955
- DOI: 10.1038/nature22363
Structure of the full-length glucagon class B G-protein-coupled receptor
Abstract
The human glucagon receptor, GCGR, belongs to the class B G-protein-coupled receptor family and plays a key role in glucose homeostasis and the pathophysiology of type 2 diabetes. Here we report the 3.0 Å crystal structure of full-length GCGR containing both the extracellular domain and transmembrane domain in an inactive conformation. The two domains are connected by a 12-residue segment termed the stalk, which adopts a β-strand conformation, instead of forming an α-helix as observed in the previously solved structure of the GCGR transmembrane domain. The first extracellular loop exhibits a β-hairpin conformation and interacts with the stalk to form a compact β-sheet structure. Hydrogen-deuterium exchange, disulfide crosslinking and molecular dynamics studies suggest that the stalk and the first extracellular loop have critical roles in modulating peptide ligand binding and receptor activation. These insights into the full-length GCGR structure deepen our understanding of the signalling mechanisms of class B G-protein-coupled receptors.
Figures








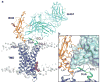




Comment in
-
A sugar high in structural biology.Nature. 2017 Jun 7;546(7657):186. doi: 10.1038/546186a. Nature. 2017. PMID: 28593982 No abstract available.
-
Newly characterized crystal structures: further insights into the architecture of GPCRs.Sci China Life Sci. 2018 May;61(5):593-596. doi: 10.1007/s11427-017-9159-6. Epub 2017 Dec 25. Sci China Life Sci. 2018. PMID: 29285710 No abstract available.
References
-
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. - PubMed
-
- Finan B, et al. Chemical Hybridization of Glucagon and Thyroid Hormone Optimizes Therapeutic Impact for Metabolic Disease. Cell. 2016;167:843–857. - PubMed
-
- Parthier C, Reedtz-Runge S, Rudolph R, Stubbs MT. Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem Sci. 2009;34:303–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

