Functional Equivalence of the SOX2 and SOX3 Transcription Factors in the Developing Mouse Brain and Testes
- PMID: 28515211
- PMCID: PMC5500146
- DOI: 10.1534/genetics.117.202549
Functional Equivalence of the SOX2 and SOX3 Transcription Factors in the Developing Mouse Brain and Testes
Abstract
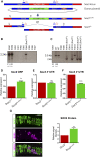
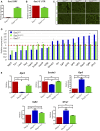
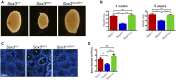
Gene duplication provides spare genetic material that evolution can craft into new functions. Sox2 and Sox3 are evolutionarily related genes with overlapping and unique sites of expression during embryogenesis. It is currently unclear whether SOX2 and SOX3 have identical or different functions. Here, we use CRISPR/Cas9-assisted mutagenesis to perform a gene-swap, replacing the Sox3 ORF with the Sox2 ORF to investigate their functional equivalence in the brain and testes. We show that increased expression of SOX2 can functionally replace SOX3 in the development of the infundibular recess/ventral diencephalon, and largely rescues pituitary gland defects that occur in Sox3 null mice. We also show that ectopic expression of SOX2 in the testes functionally rescues the spermatogenic defect of Sox3 null mice, and restores gene expression to near normal levels. Together, these in vivo data provide strong evidence that SOX2 and SOX3 proteins are functionally equivalent.
Keywords: CRISPR/CAS9 mutagenesis; SOXB1 genes; gene swap.
Copyright © 2017 by the Genetics Society of America.
Figures

References
-
- Acampora D., Annino A., Puelles E., Alfano I., Tuorto F., et al. , 2003. OTX1 compensates for OTX2 requirement in regionalisation of anterior neuroectoderm. Gene Expr. Patterns GEP 3: 497–501. - PubMed
-
- Barbaric I., Miller G., Dear T. N., 2007. Appearances can be deceiving: phenotypes of knockout mice. Brief. Funct. Genomics Proteomics 6: 91–103. - PubMed
-
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57: 289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

