A Diverse Range of Novel RNA Viruses in Geographically Distinct Honey Bee Populations
- PMID: 28515299
- PMCID: PMC5533899
- DOI: 10.1128/JVI.00158-17
A Diverse Range of Novel RNA Viruses in Geographically Distinct Honey Bee Populations
Abstract
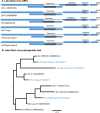
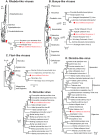
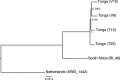
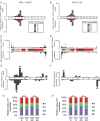
Understanding the diversity and consequences of viruses present in honey bees is critical for maintaining pollinator health and managing the spread of disease. The viral landscape of honey bees (Apis mellifera) has changed dramatically since the emergence of the parasitic mite Varroa destructor, which increased the spread of virulent variants of viruses such as deformed wing virus. Previous genomic studies have focused on colonies suffering from infections by Varroa and virulent viruses, which could mask other viral species present in honey bees, resulting in a distorted view of viral diversity. To capture the viral diversity within colonies that are exposed to mites but do not suffer the ultimate consequences of the infestation, we examined populations of honey bees that have evolved naturally or have been selected for resistance to Varroa This analysis revealed seven novel viruses isolated from honey bees sampled globally, including the first identification of negative-sense RNA viruses in honey bees. Notably, two rhabdoviruses were present in three geographically diverse locations and were also present in Varroa mites parasitizing the bees. To characterize the antiviral response, we performed deep sequencing of small RNA populations in honey bees and mites. This provided evidence of a Dicer-mediated immune response in honey bees, while the viral small RNA profile in Varroa mites was novel and distinct from the response observed in bees. Overall, we show that viral diversity in honey bee colonies is greater than previously thought, which encourages additional studies of the bee virome on a global scale and which may ultimately improve disease management.IMPORTANCE Honey bee populations have become increasingly susceptible to colony losses due to pathogenic viruses spread by parasitic Varroa mites. To date, 24 viruses have been described in honey bees, with most belonging to the order Picornavirales Collapsing Varroa-infected colonies are often overwhelmed with high levels of picornaviruses. To examine the underlying viral diversity in honey bees, we employed viral metatranscriptomics analyses on three geographically diverse Varroa-resistant populations from Europe, Africa, and the Pacific. We describe seven novel viruses from a range of diverse viral families, including two viruses that are present in all three locations. In honey bees, small RNA sequences indicate that these viruses are processed by Dicer and the RNA interference pathway, whereas Varroa mites produce strikingly novel small RNA patterns. This work increases the number and diversity of known honey bee viruses and will ultimately contribute to improved disease management in our most important agricultural pollinator.
Keywords: RNA interference; arthropod vectors; insect viruses; metagenomics; negative-strand RNA virus; phylogenetic analysis; plus-strand RNA virus.
Copyright © 2017 Remnant et al.
Figures

Similar articles
-
RNA metagenomics revealed insights into the viromes of honey bees (Apis mellifera) and Varroa mites (Varroa destructor) in Taiwan.J Invertebr Pathol. 2025 Jul;211:108341. doi: 10.1016/j.jip.2025.108341. Epub 2025 Apr 18. J Invertebr Pathol. 2025. PMID: 40254251
-
Next-generation sequence data demonstrate several pathogenic bee viruses in Middle East and African honey bee subspecies (Apis mellifera syriaca, Apis mellifera intermissa) as well as their cohabiting pathogenic mites (Varroa destructor).Virus Genes. 2018 Oct;54(5):694-705. doi: 10.1007/s11262-018-1593-9. Epub 2018 Aug 16. Virus Genes. 2018. PMID: 30116966
-
Tolerance of Honey Bees to Varroa Mite in the Absence of Deformed Wing Virus.Viruses. 2020 May 23;12(5):575. doi: 10.3390/v12050575. Viruses. 2020. PMID: 32456246 Free PMC article.
-
Understanding the Enemy: A Review of the Genetics, Behavior and Chemical Ecology of Varroa destructor, the Parasitic Mite of Apis mellifera.J Insect Sci. 2022 Jan 1;22(1):18. doi: 10.1093/jisesa/ieab101. J Insect Sci. 2022. PMID: 35137134 Free PMC article. Review.
-
Varroa destructor: A Complex Parasite, Crippling Honey Bees Worldwide.Trends Parasitol. 2020 Jul;36(7):592-606. doi: 10.1016/j.pt.2020.04.004. Epub 2020 May 23. Trends Parasitol. 2020. PMID: 32456963 Review.
Cited by
-
Unmapped RNA Virus Diversity in Termites and their Symbionts.Viruses. 2020 Oct 9;12(10):1145. doi: 10.3390/v12101145. Viruses. 2020. PMID: 33050289 Free PMC article.
-
Bee Viruses: Routes of Infection in Hymenoptera.Front Microbiol. 2020 May 28;11:943. doi: 10.3389/fmicb.2020.00943. eCollection 2020. Front Microbiol. 2020. PMID: 32547504 Free PMC article. Review.
-
Honey Bee and Bumble Bee Antiviral Defense.Viruses. 2018 Jul 27;10(8):395. doi: 10.3390/v10080395. Viruses. 2018. PMID: 30060518 Free PMC article. Review.
-
Virome Analysis of Aphid Populations That Infest the Barley Field: The Discovery of Two Novel Groups of Nege/Kita-Like Viruses and Other Novel RNA Viruses.Front Microbiol. 2020 Apr 7;11:509. doi: 10.3389/fmicb.2020.00509. eCollection 2020. Front Microbiol. 2020. PMID: 32318034 Free PMC article.
-
The First Non-LRV RNA Virus in Leishmania.Viruses. 2020 Feb 2;12(2):168. doi: 10.3390/v12020168. Viruses. 2020. PMID: 32024293 Free PMC article.
References
-
- Ellis JD, Munn PA. 2005. The worldwide health status of honey bees. Bee World 86:88–101. doi:10.1080/0005772X.2005.11417323. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

