Ligand chain length drives activation of lipid G protein-coupled receptors
- PMID: 28515494
- PMCID: PMC5435731
- DOI: 10.1038/s41598-017-02104-5
Ligand chain length drives activation of lipid G protein-coupled receptors
Abstract
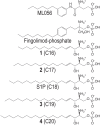
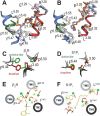
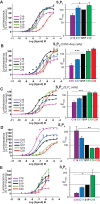
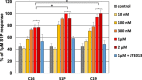
Sphingosine-1-phosphate (S1P) is a lipid mediator that can activate five cell membrane G protein-coupled receptors (GPCRs) which carry a variety of essential functions and are promising drug targets. S1P is composed of a polar zwitterionic head-group and a hydrophobic alkyl chain. This implies an activation mechanism of its cognate receptor that must be significantly different from what is known for prototypical GPCRs (ie receptor to small hydrophilic ligands). Here we aim to identify the structural features responsible for S1P agonism by combining molecular dynamics simulations and functional assays using S1P analogs of different alkyl chain lengths. We propose that high affinity binding involves polar interactions between the lipid head-group and receptor side chains while activation is due to hydrophobic interactions between the lipid tail and residues in a distinct binding site. We observe that ligand efficacy is directly related to alkyl chain length but also varies with receptor subtypes in correlation with the size of this binding pocket. Integrating experimental and computational data, we propose an activation mechanism for the S1P receptors involving agonist-induced conformational events that are conserved throughout class A GPCRs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

