Different Somatic Hypermutation Levels among Antibody Subclasses Disclosed by a New Next-Generation Sequencing-Based Antibody Repertoire Analysis
- PMID: 28515723
- PMCID: PMC5413556
- DOI: 10.3389/fimmu.2017.00389
Different Somatic Hypermutation Levels among Antibody Subclasses Disclosed by a New Next-Generation Sequencing-Based Antibody Repertoire Analysis
Abstract
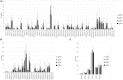
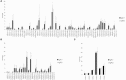
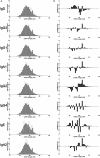
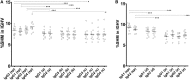
A diverse antibody repertoire is primarily generated by the rearrangement of V, D, and J genes and subsequent somatic hypermutation (SHM). Class-switch recombination (CSR) produces various isotypes and subclasses with different functional properties. Although antibody isotypes and subclasses are considered to be produced by both direct and sequential CSR, it is still not fully understood how SHMs accumulate during the process in which antibody subclasses are generated. Here, we developed a new next-generation sequencing (NGS)-based antibody repertoire analysis capable of identifying all antibody isotype and subclass genes and used it to examine the peripheral blood mononuclear cells of 12 healthy individuals. Using a total of 5,480,040 sequences, we compared percentage frequency of variable (V), junctional (J) sequence, and a combination of V and J, diversity, length, and amino acid compositions of CDR3, SHM, and shared clones in the IgM, IgD, IgG3, IgG1, IgG2, IgG4, IgA1, IgE, and IgA2 genes. The usage and diversity were similar among the immunoglobulin (Ig) subclasses. Clonally related sequences sharing identical V, D, J, and CDR3 amino acid sequences were frequently found within multiple Ig subclasses, especially between IgG1 and IgG2 or IgA1 and IgA2. SHM occurred most frequently in IgG4, while IgG3 genes were the least mutated among all IgG subclasses. The shared clones had almost the same SHM levels among Ig subclasses, while subclass-specific clones had different levels of SHM dependent on the genomic location. Given the sequential CSR, these results suggest that CSR occurs sequentially over multiple subclasses in the order corresponding to the genomic location of IGHCs, but CSR is likely to occur more quickly than SHMs accumulate within Ig genes under physiological conditions. NGS-based antibody repertoire analysis should provide critical information on how various antibodies are generated in the immune system.
Keywords: antibody; class switch recombination; repertoire; sequencing; somatic hypermutation.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

