Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7
- PMID: 28515784
- PMCID: PMC5433082
- DOI: 10.1186/s13068-017-0806-0
Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7
Abstract
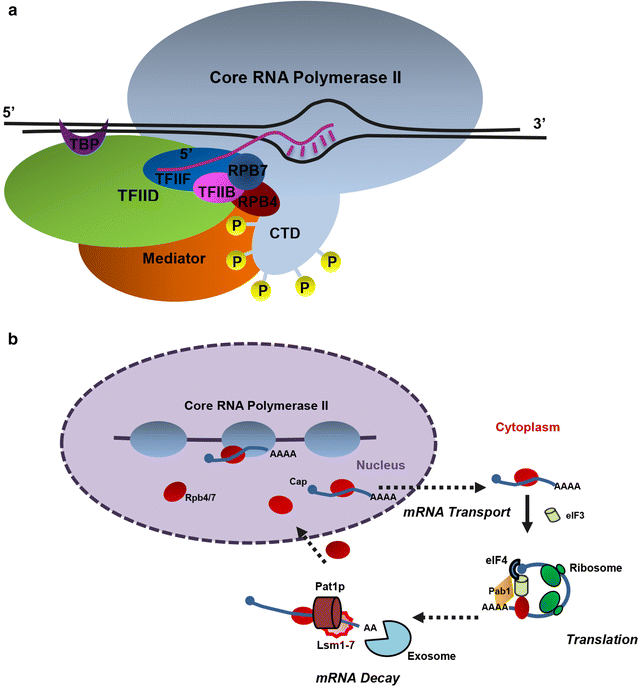
Background: Classical strain engineering methods often have limitations in altering multigenetic cellular phenotypes. Here we try to improve Saccharomyces cerevisiae ethanol tolerance and productivity by reprogramming its transcription profile through rewiring its key transcription component RNA polymerase II (RNAP II), which plays a central role in synthesizing mRNAs. This is the first report on using directed evolution method to engineer RNAP II to alter S. cerevisiae strain phenotypes.
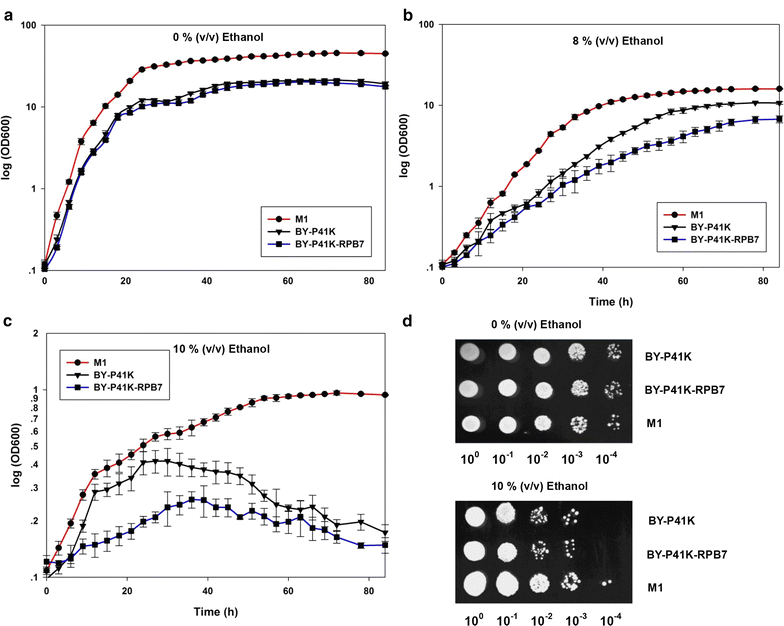
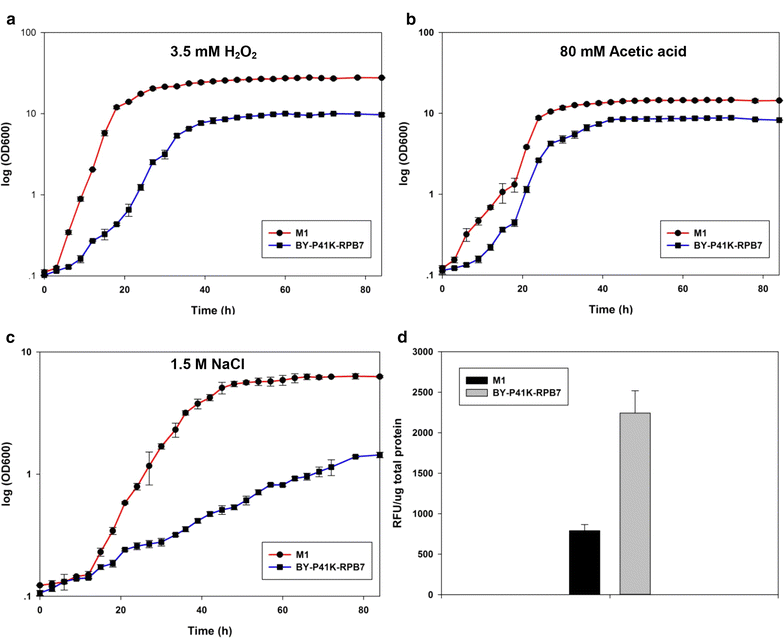
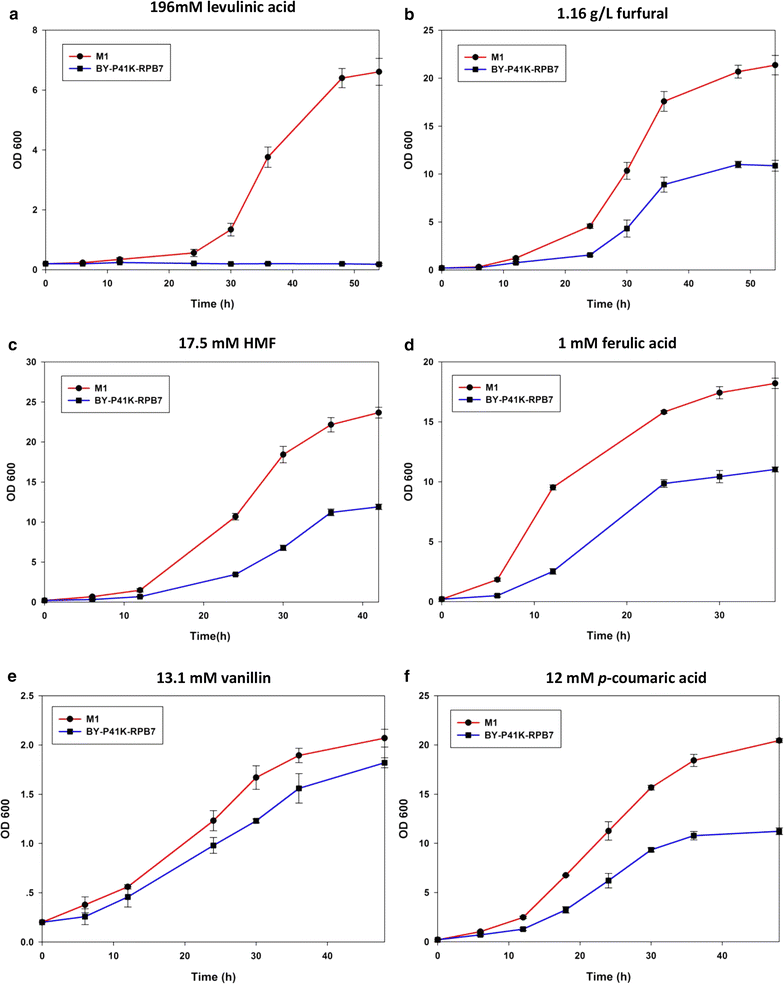
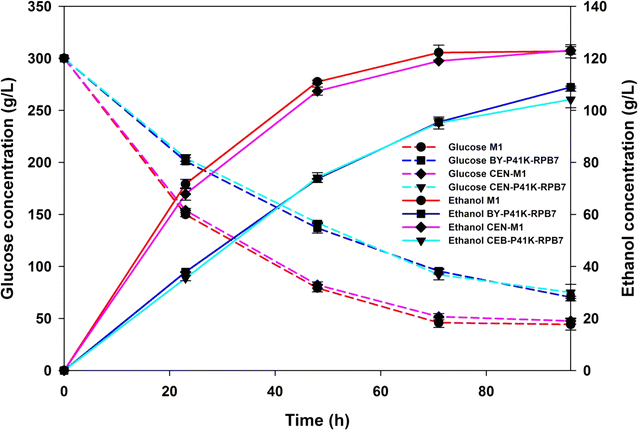
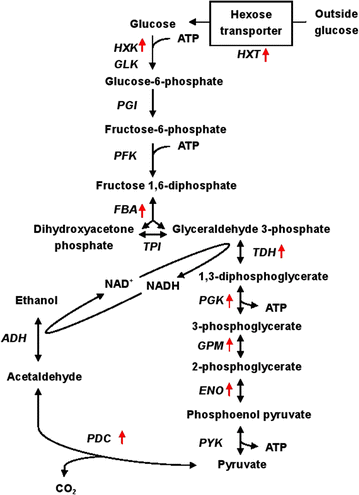
Results: Error-prone PCR was employed to engineer the subunit Rpb7 of RNAP II to improve yeast ethanol tolerance and production. Based on previous studies and the presumption that improved ethanol resistance would lead to enhanced ethanol production, we first isolated variant M1 with much improved resistance towards 8 and 10% ethanol. The ethanol titers of M1 was ~122 g/L (96.58% of the theoretical yield) under laboratory very high gravity (VHG) fermentation, 40% increase as compared to the control. DNA microarray assay showed that 369 genes had differential expression in M1 after 12 h VHG fermentation, which are involved in glycolysis, alcoholic fermentation, oxidative stress response, etc.
Conclusions: This is the first study to demonstrate the possibility of engineering eukaryotic RNAP to alter global transcription profile and improve strain phenotypes. Targeting subunit Rpb7 of RNAP II was able to bring differential expression in hundreds of genes in S. cerevisiae, which finally led to improvement in yeast ethanol tolerance and production.
Keywords: Ethanol productivity; Ethanol titers; Ethanol tolerance; Global transcription machinery engineering (gTME); Oxidative tolerance; RNA polymerase II; Subunit Rpb7; Transcriptional engineering; VHG fermentation.
Figures

References
-
- Ansanay-Galeote V, Blondin B, Dequin S, Sablayrolles JM. Stress effect of ethanol on fermentation kinetics by stationary-phase cells of Saccharomyces cerevisiae. Biotechnol Lett. 2001;23:677–681. doi: 10.1023/A:1010396232420. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

