A computational approach for the functional classification of the epigenome
- PMID: 28515787
- PMCID: PMC5433140
- DOI: 10.1186/s13072-017-0131-7
A computational approach for the functional classification of the epigenome
Abstract
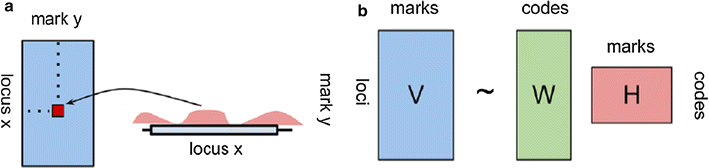
Background: In the last decade, advanced functional genomics approaches and deep sequencing have allowed large-scale mapping of histone modifications and other epigenetic marks, highlighting functional relationships between chromatin organization and genome function. Here, we propose a novel approach to explore functional interactions between different epigenetic modifications and extract combinatorial profiles that can be used to annotate the chromatin in a finite number of functional classes. Our method is based on non-negative matrix factorization (NMF), an unsupervised learning technique originally employed to decompose high-dimensional data in a reduced number of meaningful patterns. We applied the NMF algorithm to a set of different epigenetic marks, consisting of ChIP-seq assays for multiple histone modifications, Pol II binding and chromatin accessibility assays from human H1 cells.
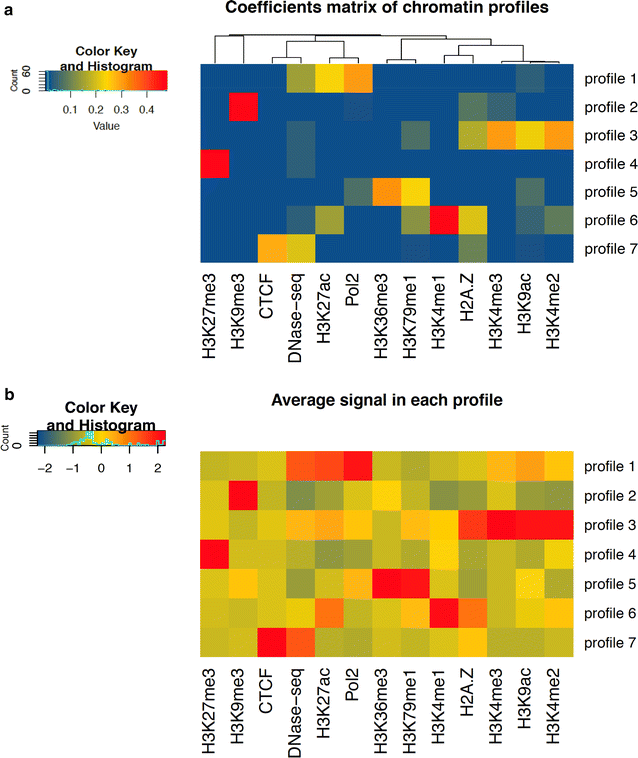
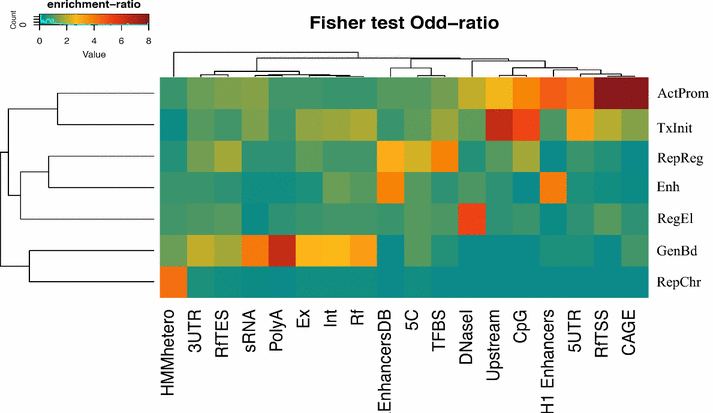
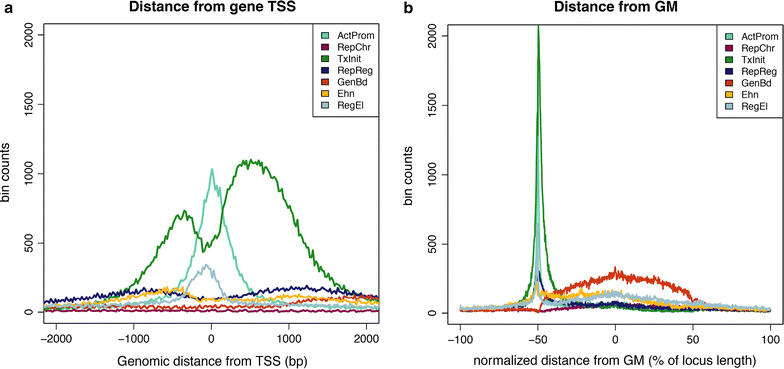
Results: We identified a number of chromatin profiles that contain functional information and are biologically interpretable. We also observe that epigenetic profiles are characterized by specific genomic contexts and show significant association with distinct genomic features. Moreover, analysis of RNA-seq data reveals that distinct chromatin signatures correlate with the level of gene expression.
Conclusions: Overall, our study highlights the utility of NMF in studying functional relationships between different epigenetic modifications and may provide new biological insights for the interpretation of the chromatin dynamics.
Keywords: Chromatin profiles; Epigenetic mark combinations; NMF.
Figures

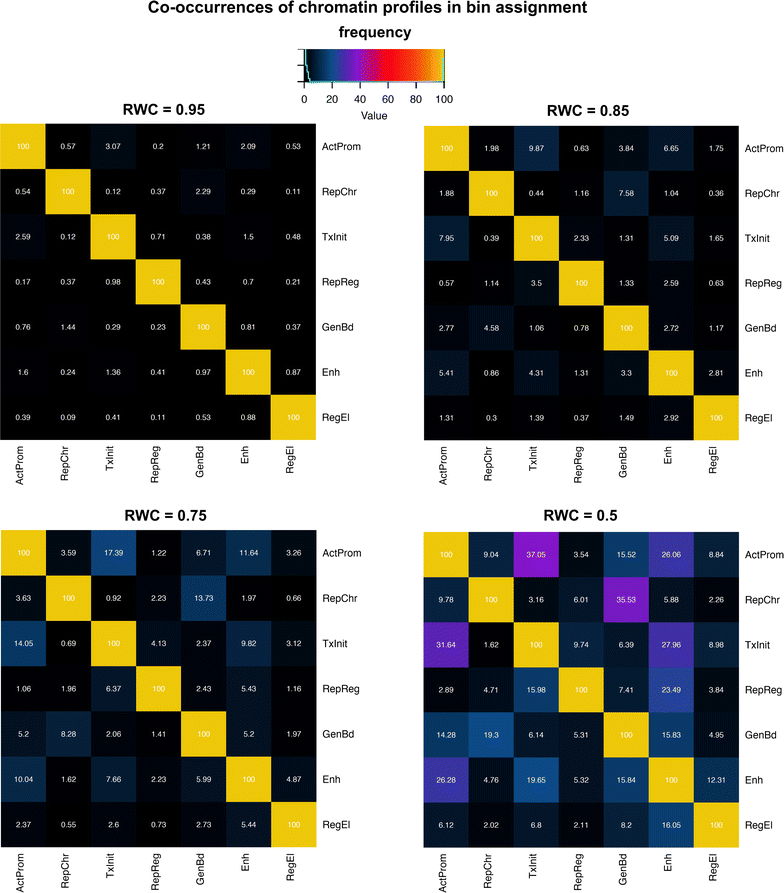
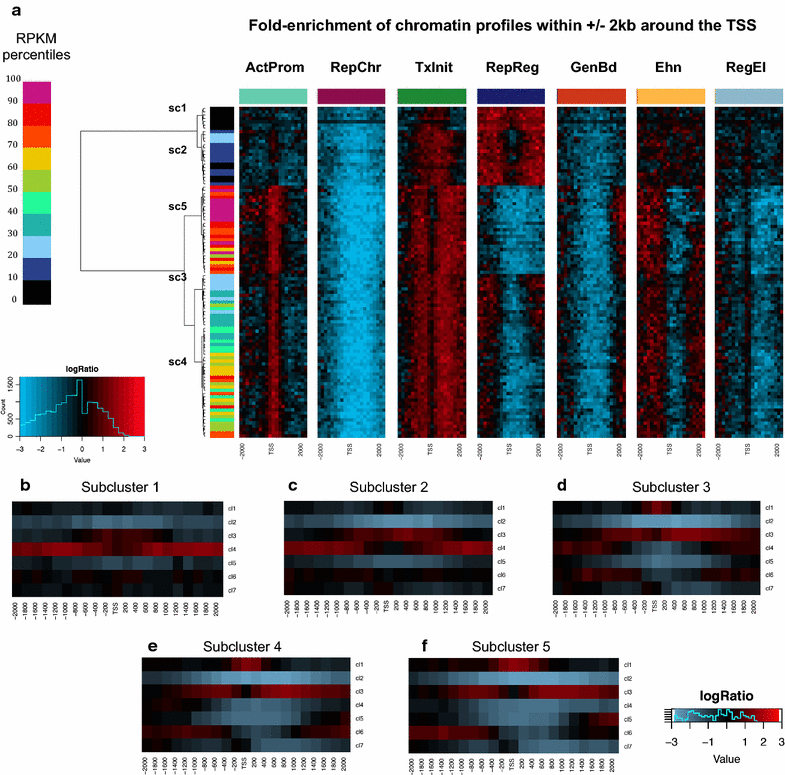
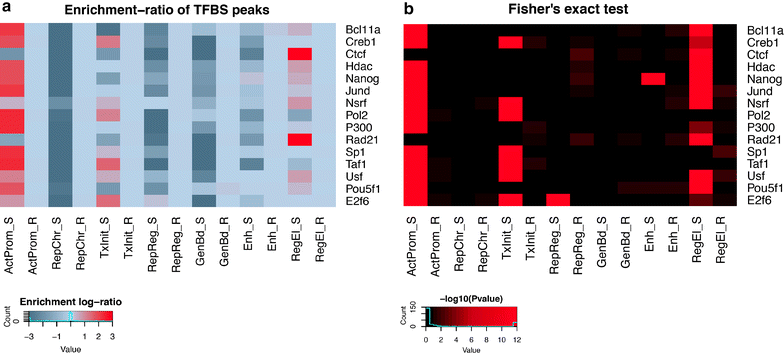
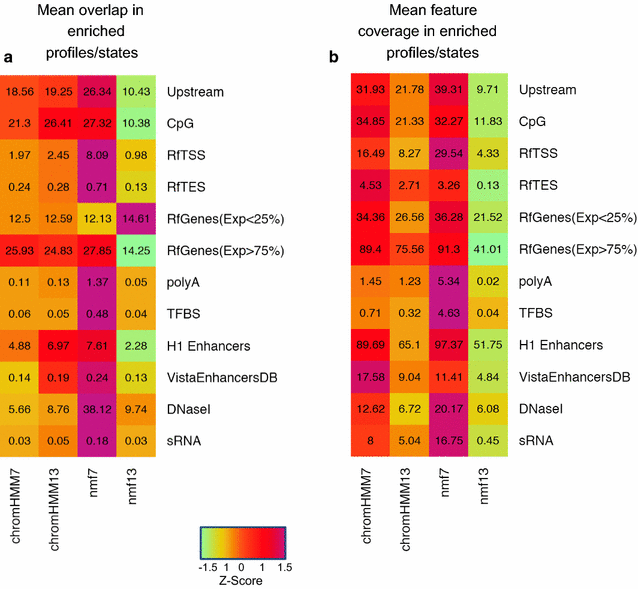

References
-
- The-Encode-Project-Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004; 306(5696):636–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

