Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer
- PMID: 28515797
- PMCID: PMC5433015
- DOI: 10.1186/s13148-017-0351-5
Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer
Abstract
Background: Colonoscopy is currently widely accepted as the gold standard for detection of colorectal cancer (CRC) providing detection of up to 95% of pre-cancerous lesions during the procedure. However, certain limitations exist in most countries including cost and access to the procedure. Moreover, colonoscopy is an invasive technique with risk inherent to the endoscopic procedure. For this reason, alternative screening tests, in particular, fecal occult blood-based tests, have been widely adopted for frontline screening. Limited compliance to colonoscopy and fecal screening approaches has prompted research on blood-based tests as an alternative approach to identifying individuals at risk who could then be referred for colonoscopy. Increased total levels of nucleosomes in the blood have been associated with tumor burden and malignancy progression. Here, we report for the first time, CRC-associated epigenetic profiles of circulating cell-free nucleosomes (cf-nucleosomes).
Methods: Levels of 12 epigenetic cf-nucleosome epitopes were measured in the sera of 58 individuals referred for endoscopic screening for CRC.
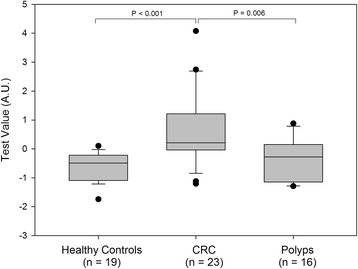
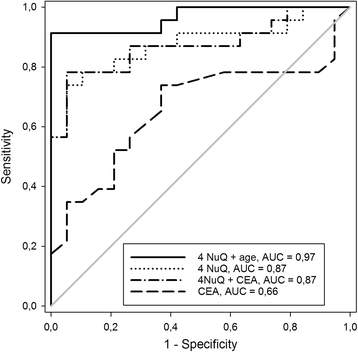
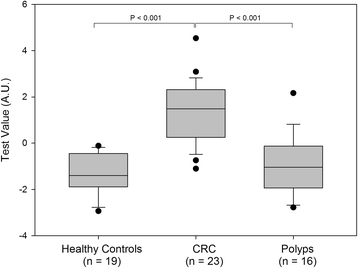
Results: Multivariate analysis defined an age-adjusted panel of four cf-nucleosomes that provided an AUC of 0.97 for the discrimination of CRC from healthy controls with high sensitivity at early stages (sensitivity of 75 and 86 at 90% specificity for stages I and II, respectively). A second combination of four cf-nucleosome biomarkers provided an AUC of 0.72 for the discrimination of polyps from the healthy group.
Conclusions: This study suggests that a combination of different cf-nucleosome structures analyzed in serum samples by a simple ELISA is a promising approach to identify patients at risk of CRC.
Keywords: Blood-based screening test; Colorectal cancer; Epigenetics; Nucleosomes.
Figures


References
-
- Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637-49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

