Polysome Fractionation to Analyze mRNA Distribution Profiles
- PMID: 28516123
- PMCID: PMC5431591
- DOI: 10.21769/BioProtoc.2126
Polysome Fractionation to Analyze mRNA Distribution Profiles
Erratum in
-
Correction: Polysome Fractionation to Analyze mRNA Distribution Profiles.Bio Protoc. 2019 Feb 5;9(3):e3176. doi: 10.21769/BioProtoc.3176. Bio Protoc. 2019. PMID: 31187050 Free PMC article.
Abstract
Eukaryotic cells adapt to changes in external or internal signals by precisely modulating the expression of specific gene products. The expression of protein-coding genes is controlled at the transcriptional and post-transcriptional levels. Among the latter steps, the regulation of translation is particularly important in cellular processes that require rapid changes in protein expression patterns. The translational efficiency of mRNAs is altered by RNA-binding proteins (RBPs) and noncoding (nc)RNAs such as microRNAs (Panda et al., 2014a and 2014b; Abdelmohsen et al., 2014). The impact of factors that regulate selective mRNA translation is a critical question in RNA biology. Polyribosome (polysome) fractionation analysis is a powerful method to assess the association of ribosomes with a given mRNA. It provides valuable information about the translational status of that mRNA, depending on the number of ribosomes with which they are associated, and identifies mRNAs that are not translated (Panda et al., 2016). mRNAs associated with many ribosomes form large polysomes that are predicted to be actively translated, while mRNAs associated with few or no ribosomes are expected to be translated poorly if at all. In sum, polysome fractionation analysis allows the direct determination of translation efficiencies at the level of the whole transcriptome as well as individual mRNAs.
Keywords: Fractionation; Polysomes; Protein synthesis; RT-qPCR; Ribosome; Sucrose gradient; mRNA translation.
Figures
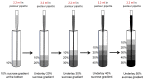
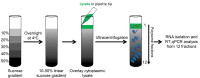
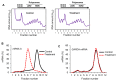
References
-
- Panda A. C., Abdelmohsen K., Martindale J. L., Di Germanio C., Yang X., Grammatikakis I., Noh J. H., Zhang Y., Lehrmann E., Dudekula D. B., De S., Becker K. G., White E. J., Wilson G. M., de Cabo R. and Gorospe M.(2016). Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res 44(5): 2393-2408. - PMC - PubMed
-
- Panda A. C., Abdelmohsen K., Yoon J. H., Martindale J. L., Yang X., Curtis J., Mercken E. M., Chenette D. M., Zhang Y., Schneider R. J., Becker K. G., de Cabo R. and Gorospe M.(2014). RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol Cell Biol 34(16): 3106-3119. - PMC - PubMed
-
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.(2011). Global quantification of mammalian gene expression control. Nature 473; 337-342. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

