An optimized strategy to measure protein stability highlights differences between cold and hot unfolded states
- PMID: 28516908
- PMCID: PMC5454340
- DOI: 10.1038/ncomms15428
An optimized strategy to measure protein stability highlights differences between cold and hot unfolded states
Abstract
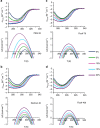
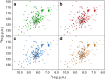
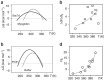
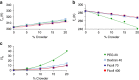
Macromolecular crowding ought to stabilize folded forms of proteins, through an excluded volume effect. This explanation has been questioned and observed effects attributed to weak interactions with other cell components. Here we show conclusively that protein stability is affected by volume exclusion and that the effect is more pronounced when the crowder's size is closer to that of the protein under study. Accurate evaluation of the volume exclusion effect is made possible by the choice of yeast frataxin, a protein that undergoes cold denaturation above zero degrees, because the unfolded form at low temperature is more expanded than the corresponding one at high temperature. To achieve optimum sensitivity to changes in stability we introduce an empirical parameter derived from the stability curve. The large effect of PEG 20 on cold denaturation can be explained by a change in water activity, according to Privalov's interpretation of cold denaturation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Zimmerman S. B. & Trach S. O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 222, 599–620 (1991). - PubMed
-
- Politou A. & Temussi P. A. Revisiting a dogma: the effect of volume exclusion in molecular crowding. Curr. Opin. Struct. Biol. 30, 1–6 (2015). - PubMed
-
- Smith A. E., Zhang Z., Pielak G. J. & Li C. NMR studies of protein folding and binding in cells and cell-like environments. Curr. Opin. Struct. Biol. 30, 7–16 (2015). - PubMed
-
- Oshima H. & Kinoshita M. Effects of sugars on the thermal stability of a protein. J. Chem. Phys. 138, 245101 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

