Transcriptomic analysis of salt stress responsive genes in Rhazya stricta
- PMID: 28520766
- PMCID: PMC5433744
- DOI: 10.1371/journal.pone.0177589
Transcriptomic analysis of salt stress responsive genes in Rhazya stricta
Abstract
Rhazya stricta is an evergreen shrub that is widely distributed across Western and South Asia, and like many other members of the Apocynaceae produces monoterpene indole alkaloids that have anti-cancer properties. This species is adapted to very harsh desert conditions making it an excellent system for studying tolerance to high temperatures and salinity. RNA-Seq analysis was performed on R. stricta exposed to severe salt stress (500 mM NaCl) across four time intervals (0, 2, 12 and 24 h) to examine mechanisms of salt tolerance. A large number of transcripts including genes encoding tetrapyrroles and pentatricopeptide repeat (PPR) proteins were regulated only after 12 h of stress of seedlings grown in controlled greenhouse conditions. Mechanisms of salt tolerance in R. stricta may involve the upregulation of genes encoding chaperone protein Dnaj6, UDP-glucosyl transferase 85a2, protein transparent testa 12 and respiratory burst oxidase homolog protein b. Many of the highly-expressed genes act on protecting protein folding during salt stress and the production of flavonoids, key secondary metabolites in stress tolerance. Other regulated genes encode enzymes in the porphyrin and chlorophyll metabolic pathway with important roles during plant growth, photosynthesis, hormone signaling and abiotic responses. Heme biosynthesis in R. stricta leaves might add to the level of salt stress tolerance by maintaining appropriate levels of photosynthesis and normal plant growth as well as by the participation in reactive oxygen species (ROS) production under stress. We speculate that the high expression levels of PPR genes may be dependent on expression levels of their targeted editing genes. Although the results of PPR gene family indicated regulation of a large number of transcripts under salt stress, PPR actions were independent of the salt stress because their RNA editing patterns were unchanged.
Conflict of interest statement
Figures

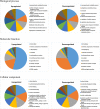
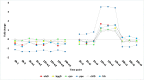
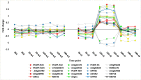
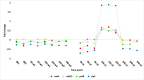
References
-
- Xiong L, Zhu JK. Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol Plant. 2001; 112:152–166. - PubMed
-
- Zhu JK Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003; 6:441–445. - PubMed
-
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sunchez-Serrano JJ, Schmelz EA, et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 2007; 19:1665–1681. doi: 10.1105/tpc.106.048041 - DOI - PMC - PubMed
-
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Ann Rev Plant Biol. 2010; 61:651–679. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

