Genomic epidemiology of global VIM-producing Enterobacteriaceae
- PMID: 28520983
- PMCID: PMC5890710
- DOI: 10.1093/jac/dkx148
Genomic epidemiology of global VIM-producing Enterobacteriaceae
Abstract
Background: International data on the molecular epidemiology of Enterobacteriaceae with VIM carbapenemases are limited.
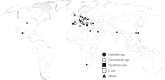
Methods: We performed short read (Illumina) WGS on a global collection of 89 VIM-producing clinical Enterobacteriaceae (2008-14).
Results: VIM-producing (11 varieties within 21 different integrons) isolates were mostly obtained from Europe. Certain integrons with bla VIM were specific to a country in different species and clonal complexes (CCs) (In 87 , In 624 , In 916 and In 1323 ), while others had spread globally among various Enterobacteriaceae species (In 110 and In 1209 ). Klebsiella pneumoniae was the most common species ( n = 45); CC147 from Greece was the most prevalent clone and contained In 590 -like integrons with four different bla VIM s. Enterobacter cloacae complex was the second most common species and mainly consisted of Enterobacter hormaechei ( Enterobacter xiangfangensis , subsp. steigerwaltii and Hoffmann cluster III). CC200 (from Croatia and Turkey), CC114 (Croatia, Greece, Italy and the USA) and CC78 (from Greece, Italy and Spain) containing bla VIM-1 were the most common clones among the E. cloacae complex.
Conclusions: This study highlights the importance of surveillance programmes using the latest molecular techniques in providing insight into the characteristics and global distribution of Enterobacteriaceae with bla VIM s.
© The Author 2017. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Albiger B, Glasner C, Struelens MJ. et al. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill 2015; 20: pii=30062. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

