The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders
- PMID: 28521612
- PMCID: PMC5446072
- DOI: 10.1080/15548627.2017.1291470
The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders
Abstract
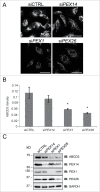
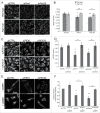
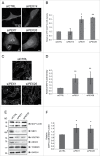
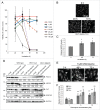
Peroxisome biogenesis disorders (PBDs) are metabolic disorders caused by the loss of peroxisomes. The majority of PBDs result from mutation in one of 3 genes that encode for the peroxisomal AAA ATPase complex (AAA-complex) required for cycling PEX5 for peroxisomal matrix protein import. Mutations in these genes are thought to result in a defect in peroxisome assembly by preventing the import of matrix proteins. However, we show here that loss of the AAA-complex does not prevent matrix protein import, but instead causes an upregulation of peroxisome degradation by macroautophagy, or pexophagy. The loss of AAA-complex function in cells results in the accumulation of ubiquitinated PEX5 on the peroxisomal membrane that signals pexophagy. Inhibiting autophagy by genetic or pharmacological approaches rescues peroxisome number, protein import and function. Our findings suggest that the peroxisomal AAA-complex is required for peroxisome quality control, whereas its absence results in the selective degradation of the peroxisome. Thus the loss of peroxisomes in PBD patients with mutations in their peroxisomal AAA-complex is a result of increased pexophagy. Our study also provides a framework for the development of novel therapeutic treatments for PBDs.
Keywords: AAA ATPase complex; PEX1; PEX26; PEX5; Zellweger spectrum disorder; autophagy; peroxisome biogenesis disorder; peroxisomes; pexophagy; selective autophagy.
Figures



Similar articles
-
Pexophagy is responsible for 65% of cases of peroxisome biogenesis disorders.Autophagy. 2017 May 4;13(5):991-994. doi: 10.1080/15548627.2017.1291480. Epub 2017 Feb 28. Autophagy. 2017. PMID: 28318378 Free PMC article. Review.
-
A pex1 missense mutation improves peroxisome function in a subset of Arabidopsis pex6 mutants without restoring PEX5 recycling.Proc Natl Acad Sci U S A. 2018 Apr 3;115(14):E3163-E3172. doi: 10.1073/pnas.1721279115. Epub 2018 Mar 19. Proc Natl Acad Sci U S A. 2018. PMID: 29555730 Free PMC article.
-
The Peroxisomal PTS1-Import Defect of PEX1- Deficient Cells Is Independent of Pexophagy in Saccharomyces cerevisiae.Int J Mol Sci. 2020 Jan 29;21(3):867. doi: 10.3390/ijms21030867. Int J Mol Sci. 2020. PMID: 32013259 Free PMC article.
-
PEX13 prevents pexophagy by regulating ubiquitinated PEX5 and peroxisomal ROS.Autophagy. 2023 Jun;19(6):1781-1802. doi: 10.1080/15548627.2022.2160566. Epub 2023 Jan 1. Autophagy. 2023. PMID: 36541703 Free PMC article.
-
Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease.Biochim Biophys Acta. 2012 Sep;1822(9):1337-42. doi: 10.1016/j.bbadis.2012.06.004. Epub 2012 Jun 13. Biochim Biophys Acta. 2012. PMID: 22705440 Review.
Cited by
-
Peroxisomal Stress Response and Inter-Organelle Communication in Cellular Homeostasis and Aging.Antioxidants (Basel). 2022 Jan 19;11(2):192. doi: 10.3390/antiox11020192. Antioxidants (Basel). 2022. PMID: 35204075 Free PMC article. Review.
-
Knockdown of PEX16 Induces Autophagic Degradation of Peroxisomes.Int J Mol Sci. 2021 Jul 26;22(15):7989. doi: 10.3390/ijms22157989. Int J Mol Sci. 2021. PMID: 34360754 Free PMC article.
-
Selective autophagy of intracellular organelles: recent research advances.Theranostics. 2021 Jan 1;11(1):222-256. doi: 10.7150/thno.49860. eCollection 2021. Theranostics. 2021. PMID: 33391472 Free PMC article.
-
Regulatory Mechanism of Peroxisome Number Reduction Caused by FgPex4 and FgPex22-like Deletion in Fusarium graminearum.J Fungi (Basel). 2023 Nov 6;9(11):1083. doi: 10.3390/jof9111083. J Fungi (Basel). 2023. PMID: 37998888 Free PMC article.
-
Insights into the Structure and Function of the Pex1/Pex6 AAA-ATPase in Peroxisome Homeostasis.Cells. 2022 Jun 29;11(13):2067. doi: 10.3390/cells11132067. Cells. 2022. PMID: 35805150 Free PMC article. Review.
References
-
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ [Internet] 2005; 12 Suppl 2:1542-52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16247502; http://dx.doi.org/ 10.1038/sj.cdd.4401765 - DOI - PMC - PubMed
-
- Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol [Internet] 2014; 205:435-45. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24862571; PMID: 24862571; http://dx.doi.org/ 10.1083/jcb.201402054 - DOI - PMC - PubMed
-
- Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol [Internet] 2014; 16:495-501. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24875736; PMID: 24875736; http://dx.doi.org/ 10.1038/ncb2979 - DOI - PubMed
-
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy [Internet] 2011; 7:279-96. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21189453; PMID: 21189453; http://dx.doi.org/ 10.4161/auto.7.3.14487 - DOI - PMC - PubMed
-
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron [Internet] 2015; 85:257-73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25611507; http://dx.doi.org/ 10.1016/j.neuron.2014.12.007 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
