Interpreting the functional role of a novel interaction motif in prokaryotic sodium channels
- PMID: 28522439
- PMCID: PMC5460950
- DOI: 10.1085/jgp.201611740
Interpreting the functional role of a novel interaction motif in prokaryotic sodium channels
Abstract
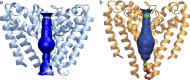
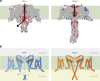
Voltage-gated sodium channels enable the translocation of sodium ions across cell membranes and play crucial roles in electrical signaling by initiating the action potential. In humans, mutations in sodium channels give rise to several neurological and cardiovascular diseases, and hence they are targets for pharmaceutical drug developments. Prokaryotic sodium channel crystal structures have provided detailed views of sodium channels, which by homology have suggested potentially important functionally related structural features in human sodium channels. A new crystal structure of a full-length prokaryotic channel, NavMs, in a conformation we proposed to represent the open, activated state, has revealed a novel interaction motif associated with channel opening. This motif is associated with disease when mutated in human sodium channels and plays an important and dynamic role in our new model for channel activation.
© 2017 Sula and Wallace.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

