A Novel Histone Crosstalk Pathway Important for Regulation of UV-Induced DNA Damage Repair in Saccharomyces cerevisiae
- PMID: 28522541
- PMCID: PMC5500138
- DOI: 10.1534/genetics.116.195735
A Novel Histone Crosstalk Pathway Important for Regulation of UV-Induced DNA Damage Repair in Saccharomyces cerevisiae
Abstract
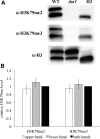
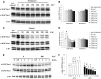
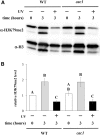
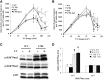
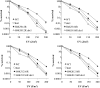
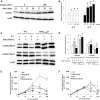
Histone post-translational modifications play vital roles in a variety of nuclear processes, including DNA repair. It has been previously shown that histone H3K79 methylation is important for the cellular response to DNA damage caused by ultraviolet (UV) radiation, with evidence that specific methylation states play distinct roles in UV repair. Here, we report that H3K79 methylation is reduced in response to UV exposure in Saccharomyces cerevisiae This reduction is specific to the dimethylated state, as trimethylation levels are minimally altered by UV exposure. Inhibition of this reduction has a deleterious effect on UV-induced sister chromatid exchange, suggesting that H3K79 dimethylation levels play a regulatory role in UV repair. Further evidence implicates an additional role for H3K79 dimethylation levels in error-free translesion synthesis, but not in UV-induced G1/S checkpoint activation or double-stranded break repair. Additionally, we find that H3K79 dimethylation levels are influenced by acetylatable lysines on the histone H4 N-terminal tail, which are hyperacetylated in response to UV exposure. Preclusion of H4 acetylation prevents UV-induced reduction of H3K79 dimethylation, and similarly has a negative effect on UV-induced sister chromatid exchange. These results point to the existence of a novel histone crosstalk pathway that is important for the regulation of UV-induced DNA damage repair.
Keywords: DNA repair; histone crosstalk; histone modification; methylation; sister chromatid exchange.
Copyright © 2017 by the Genetics Society of America.
Figures




References
-
- Bostelman L. J., Keller A. M., Albrecht A. M., Arat A., Thompson J. S., 2007. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 6: 383–395. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

