A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants
- PMID: 28522548
- PMCID: PMC5502448
- DOI: 10.1105/tpc.16.00922
A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants
Abstract
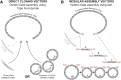
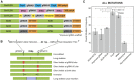
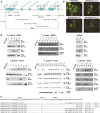
We report a comprehensive toolkit that enables targeted, specific modification of monocot and dicot genomes using a variety of genome engineering approaches. Our reagents, based on transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, are systematized for fast, modular cloning and accommodate diverse regulatory sequences to drive reagent expression. Vectors are optimized to create either single or multiple gene knockouts and large chromosomal deletions. Moreover, integration of geminivirus-based vectors enables precise gene editing through homologous recombination. Regulation of transcription is also possible. A Web-based tool streamlines vector selection and construction. One advantage of our platform is the use of the Csy-type (CRISPR system yersinia) ribonuclease 4 (Csy4) and tRNA processing enzymes to simultaneously express multiple guide RNAs (gRNAs). For example, we demonstrate targeted deletions in up to six genes by expressing 12 gRNAs from a single transcript. Csy4 and tRNA expression systems are almost twice as effective in inducing mutations as gRNAs expressed from individual RNA polymerase III promoters. Mutagenesis can be further enhanced 2.5-fold by incorporating the Trex2 exonuclease. Finally, we demonstrate that Cas9 nickases induce gene targeting at frequencies comparable to native Cas9 when they are delivered on geminivirus replicons. The reagents have been successfully validated in tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), Medicago truncatula, wheat (Triticum aestivum), and barley (Hordeum vulgare).
© 2017 American Society of Plant Biologists. All rights reserved.
Figures







References
-
- Baltes N.J., Voytas D.F. (2015). Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33: 120–131. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

