Genome Analysis of Conserved Dehydrin Motifs in Vascular Plants
- PMID: 28523013
- PMCID: PMC5415607
- DOI: 10.3389/fpls.2017.00709
Genome Analysis of Conserved Dehydrin Motifs in Vascular Plants
Abstract
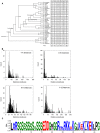
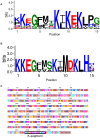
Dehydrins, a large family of abiotic stress proteins, are defined by the presence of a mostly conserved motif known as the K-segment, and may also contain two other conserved motifs known as the Y-segment and S-segment. Using the dehydrin literature, we developed a sequence motif definition of the K-segment, which we used to create a large dataset of dehydrin sequences by searching the Pfam00257 dehydrin dataset and the Phytozome 10 sequences of vascular plants. A comprehensive analysis of these sequences reveals that lysine residues are highly conserved in the K-segment, while the amino acid type is often conserved at other positions. Despite the Y-segment name, the central tyrosine is somewhat conserved, but can be substituted with two other small aromatic amino acids (phenylalanine or histidine). The S-segment contains a series of serine residues, but in some proteins is also preceded by a conserved LHR sequence. In many dehydrins containing all three of these motifs the S-segment is linked to the K-segment by a GXGGRRKK motif (where X can be any amino acid), suggesting a functional linkage between these two motifs. An analysis of the sequences shows that the dehydrin architecture and several biochemical properties (isoelectric point, molecular mass, and hydrophobicity score) are dependent on each other, and that some dehydrin architectures are overexpressed during certain abiotic stress, suggesting that they may be optimized for a specific abiotic stress while others are involved in all forms of dehydration stress (drought, cold, and salinity).
Keywords: abiotic stress; cold stress; dehydrin; drought stress; intrinsically disordered protein; motif search; phytozome; salinity.
Figures








References
-
- Ahmad P., Prasad M. N. V. (eds) (2012). Abiotic Stress Responses in Plants. New York, NY: Springer; 10.1007/978-1-4614-0634-1 - DOI
-
- Alsheikh M. K., Svensson J. T., Randall S. K. (2005). Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant Cell Environ. 28 1114–1122. 10.1111/j.1365-3040.2005.01348.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

