Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing
- PMID: 28526081
- PMCID: PMC5437632
- DOI: 10.1186/s12920-017-0271-4
Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing
Abstract
Background: The growing number of Next Generation Sequencing (NGS) tests is transforming the routine clinical diagnosis of hereditary cancers. Identifying whether a cancer is the result of an underlying disease-causing mutation in a cancer predisposition gene is not only diagnostic for a cancer predisposition syndrome, but also has significant clinical implications in the clinical management of patients and their families.
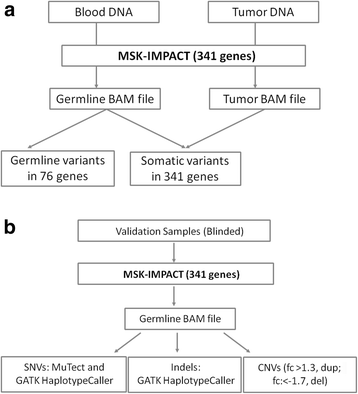
Methods: Here, we evaluated the performance of MSK-IMPACT (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets) in detecting genetic alterations in 76 genes implicated in cancer predisposition syndromes. Output from hybridization-based capture was sequenced on an Illumina HiSeq 2500. A custom analysis pipeline was used to detect single nucleotide variants (SNVs), small insertions/deletions (indels) and copy number variants (CNVs).
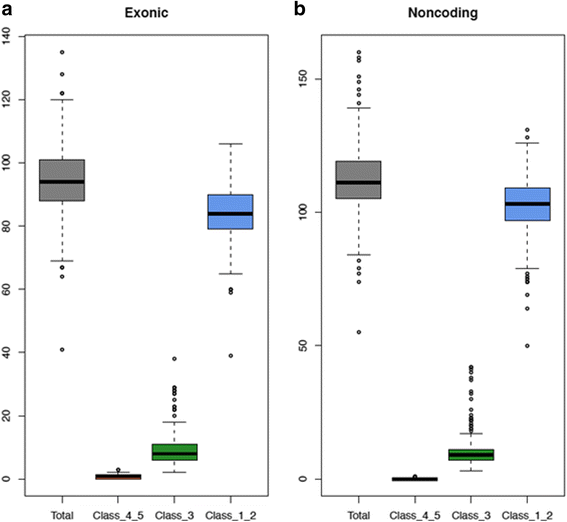
Results: MSK-IMPACT detected all germline variants in a set of 233 unique patient DNA samples, previously confirmed by previous single gene testing. Reproducibility of variant calls was demonstrated using inter- and intra- run replicates. Moreover, in 16 samples, we identified additional pathogenic mutations other than those previously identified through a traditional gene-by-gene approach, including founder mutations in BRCA1, BRCA2, CHEK2 and APC, and truncating mutations in TP53, TSC2, ATM and VHL.
Conclusions: This study highlights the importance of the NGS-based gene panel testing approach in comprehensively identifying germline variants contributing to cancer predisposition and simultaneous detection of somatic and germline alterations.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

