Adaptor proteins GIR1 and GIR2. II. Interaction with the co-repressor TOPLESS and promotion of histone deacetylation of target chromatin
- PMID: 28526412
- PMCID: PMC9423023
- DOI: 10.1016/j.bbrc.2017.05.085
Adaptor proteins GIR1 and GIR2. II. Interaction with the co-repressor TOPLESS and promotion of histone deacetylation of target chromatin
Abstract
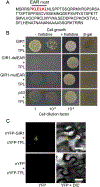
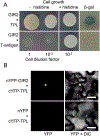
Understanding how root hair development is controlled is important for understanding of many fundamental aspects of plant biology. Previously, we identified two plant-specific adaptor proteins GIR1 and GIR2 that interact with the major regulator of root hair development GL2 and suppress formation of root hair. Here, we show that GIR1 and GIR2 also interact with the co-repressor TOPLESS (TPL). This interaction required the GIR1 protein EAR motif, and was essential for the transcriptional repressor activity of GIR1. Both GIR1 and GIR2 promoted histone hypoacetylation of their target chromatin. Potentially, GIR1 and GIR2 might may link TPL to and participate in epigenetic regulation of root hair development.
Keywords: Arabidopsis; Epigenetic regulation; GL2-Interacting proteins; Root hair development; TPL.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Adaptor proteins GIR1 and GIR2. I. Interaction with the repressor GLABRA2 and regulation of root hair development.Biochem Biophys Res Commun. 2017 Jul 1;488(3):547-553. doi: 10.1016/j.bbrc.2017.05.084. Epub 2017 May 17. Biochem Biophys Res Commun. 2017. PMID: 28526410 Free PMC article.
-
Arabidopsis ECAP Is a New Adaptor Protein that Connects JAZ Repressors with the TPR2 Co-repressor to Suppress Jasmonate-Responsive Anthocyanin Accumulation.Mol Plant. 2020 Feb 3;13(2):246-265. doi: 10.1016/j.molp.2019.10.014. Epub 2019 Nov 6. Mol Plant. 2020. PMID: 31706031
-
A chromatin link that couples cell division to root epidermis patterning in Arabidopsis.Nature. 2007 May 10;447(7141):213-7. doi: 10.1038/nature05763. Epub 2007 Apr 22. Nature. 2007. PMID: 17450124
-
Repressor for hire! The vital roles of TOPLESS-mediated transcriptional repression in plants.New Phytol. 2021 Aug;231(3):963-973. doi: 10.1111/nph.17428. Epub 2021 May 30. New Phytol. 2021. PMID: 33909309 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
-
PlantEAR: Functional Analysis Platform for Plant EAR Motif-Containing Proteins.Front Genet. 2018 Nov 30;9:590. doi: 10.3389/fgene.2018.00590. eCollection 2018. Front Genet. 2018. PMID: 30555515 Free PMC article.
-
Genome-Wide Analysis of SRNF Genes in Gossypium hirsutum Reveals the Role of GhSRNF18 in Primary Root Growth.Front Plant Sci. 2021 Sep 22;12:731834. doi: 10.3389/fpls.2021.731834. eCollection 2021. Front Plant Sci. 2021. PMID: 34630480 Free PMC article.
-
Analysis of Rice Proteins with DLN Repressor Motif/S.Int J Mol Sci. 2019 Mar 30;20(7):1600. doi: 10.3390/ijms20071600. Int J Mol Sci. 2019. PMID: 30935059 Free PMC article.
-
Genome-wide identification and expression analysis of GL2-interacting-repressor (GIR) genes during cotton fiber and fuzz development.Planta. 2021 Dec 19;255(1):23. doi: 10.1007/s00425-021-03737-7. Planta. 2021. PMID: 34923605
-
NbCycB2 represses Nbwo activity via a negative feedback loop in tobacco trichome development.J Exp Bot. 2020 Mar 25;71(6):1815-1827. doi: 10.1093/jxb/erz542. J Exp Bot. 2020. PMID: 31990970 Free PMC article.
References
-
- Gilroy S, Jones DL, Through form to function: root hair development and nutrient uptake, Trends Plant Sci. 5 (2000) 56–60. - PubMed
-
- Haling RE, Brown LK, Bengough AG, Young IM, Hallett PD, White PJ, George TS, Root hairs improve root penetration, root-soil contact, and phosphorus acquisition in soils of different strength, J. Exp. Bot 64 (2013) 3711–3721. - PubMed
-
- Ishida T, Kurata T, Okada K, Wada T, A genetic regulatory network in the development of trichomes and root hairs, Annu. Rev. Plant Biol 59 (2008) 365–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

