CONSTANS Imparts DNA Sequence Specificity to the Histone Fold NF-YB/NF-YC Dimer
- PMID: 28526714
- PMCID: PMC5502446
- DOI: 10.1105/tpc.16.00864
CONSTANS Imparts DNA Sequence Specificity to the Histone Fold NF-YB/NF-YC Dimer
Abstract
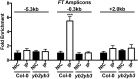
Nuclear Factor Y (NF-Y) is a heterotrimeric transcription factor that binds CCAAT elements. The NF-Y trimer is composed of a Histone Fold Domain (HFD) dimer (NF-YB/NF-YC) and NF-YA, which confers DNA sequence specificity. NF-YA shares a conserved domain with the CONSTANS, CONSTANS-LIKE, TOC1 (CCT) proteins. We show that CONSTANS (CO/B-BOX PROTEIN1 BBX1), a master flowering regulator, forms a trimer with Arabidopsis thaliana NF-YB2/NF-YC3 to efficiently bind the CORE element of the FLOWERING LOCUS T promoter. We term this complex NF-CO. Using saturation mutagenesis, electrophoretic mobility shift assays, and RNA-sequencing profiling of co, nf-yb, and nf-yc mutants, we identify CCACA elements as the core NF-CO binding site. CO physically interacts with the same HFD surface required for NF-YA association, as determined by mutations in NF-YB2 and NF-YC9, and tested in vitro and in vivo. The co-7 mutation in the CCT domain, corresponding to an NF-YA arginine directly involved in CCAAT recognition, abolishes NF-CO binding to DNA. In summary, a unifying molecular mechanism of CO function relates it to the NF-YA paradigm, as part of a trimeric complex imparting sequence specificity to HFD/DNA interactions. It is likely that members of the large CCT family participate in similar complexes with At-NF-YB and At-NF-YC, broadening HFD combinatorial possibilities in terms of trimerization, DNA binding specificities, and transcriptional regulation.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures






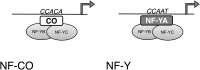
Comment in
-
CONSTANS Companion: CO Binds the NF-YB/NF-YC Dimer and Confers Sequence-Specific DNA Binding.Plant Cell. 2017 Jun;29(6):1183. doi: 10.1105/tpc.17.00465. Epub 2017 Jun 13. Plant Cell. 2017. PMID: 28611020 Free PMC article. No abstract available.
Similar articles
-
Molecular condensation of the CO/NF-YB/NF-YC/FT complex gates floral transition in Arabidopsis.EMBO J. 2025 Jan;44(1):225-250. doi: 10.1038/s44318-024-00293-0. Epub 2024 Nov 20. EMBO J. 2025. PMID: 39567828 Free PMC article.
-
Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.Plant J. 2021 Jan;105(1):49-61. doi: 10.1111/tpj.15038. Epub 2020 Nov 27. Plant J. 2021. PMID: 33098724
-
NUCLEAR FACTOR Y, Subunit A (NF-YA) Proteins Positively Regulate Flowering and Act Through FLOWERING LOCUS T.PLoS Genet. 2016 Dec 15;12(12):e1006496. doi: 10.1371/journal.pgen.1006496. eCollection 2016 Dec. PLoS Genet. 2016. PMID: 27977687 Free PMC article.
-
Plant Flowering: Imposing DNA Specificity on Histone-Fold Subunits.Trends Plant Sci. 2018 Apr;23(4):293-301. doi: 10.1016/j.tplants.2017.12.005. Epub 2018 Jan 10. Trends Plant Sci. 2018. PMID: 29331540 Review.
-
Structural determinants for NF-Y/DNA interaction at the CCAAT box.Biochim Biophys Acta Gene Regul Mech. 2017 May;1860(5):571-580. doi: 10.1016/j.bbagrm.2016.09.006. Epub 2016 Sep 25. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27677949 Review.
Cited by
-
Five NUCLEAR FACTOR-Y subunit B genes in rapeseed (Brassica napus) promote flowering and root elongation in Arabidopsis.Planta. 2022 Nov 12;256(6):115. doi: 10.1007/s00425-022-04030-x. Planta. 2022. PMID: 36371542
-
Genome-Wide Identification and Chilling Stress Analysis of the NF-Y Gene Family in Melon.Int J Mol Sci. 2023 Apr 8;24(8):6934. doi: 10.3390/ijms24086934. Int J Mol Sci. 2023. PMID: 37108097 Free PMC article.
-
Stable and dynamic gene expression patterns over diurnal and developmental timescales in Arabidopsis thaliana.New Phytol. 2025 May;246(3):1147-1162. doi: 10.1111/nph.70023. Epub 2025 Mar 20. New Phytol. 2025. PMID: 40114416 Free PMC article.
-
Molecular condensation of the CO/NF-YB/NF-YC/FT complex gates floral transition in Arabidopsis.EMBO J. 2025 Jan;44(1):225-250. doi: 10.1038/s44318-024-00293-0. Epub 2024 Nov 20. EMBO J. 2025. PMID: 39567828 Free PMC article.
-
GhCOL2 Positively Regulates Flowering by Activating the Transcription of GhHD3A in Upland Cotton (Gossypium hirsutum L.).Biochem Genet. 2025 Feb;63(1):298-314. doi: 10.1007/s10528-024-10727-3. Epub 2024 Mar 4. Biochem Genet. 2025. PMID: 38436815
References
-
- Abelenda J.A., Cruz-Oró E., Franco-Zorrilla J.M., Prat S. (2016). Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26: 872–881. - PubMed
-
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46: 462–476. - PubMed
-
- Blackman B.K., Michaels S.D. (2010). Does CONSTANS act as a transcription factor or as a co-activator? The answer may be--yes. New Phytol. 187: 1–3. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

