Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis
- PMID: 28526747
- PMCID: PMC5500816
- DOI: 10.1074/jbc.M116.774067
Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis
Abstract
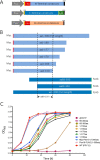
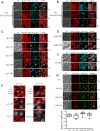
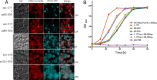
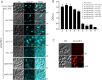
The peroxins Pex19 and Pex3 play an indispensable role in peroxisomal membrane protein (PMP) biogenesis, peroxisome division, and inheritance. Pex19 plays multiple roles in these processes, but how these functions relate to the structural organization of the Pex19 domains is unresolved. To this end, using deletion mutants, we mapped the Pex19 regions required for peroxisome biogenesis in the yeast Pichia pastoris Surprisingly, import-competent peroxisomes still formed when Pex19 domains previously believed to be required for biogenesis were deleted, although the peroxisome size was larger than that in wild-type cells. Moreover, these mutants exhibited a delay of 14-24 h in peroxisome biogenesis. The shortest functional N-terminal (NTCs) and C-terminal constructs (CTCs) were Pex19 (aa 1-150) and Pex19 (aa 89-300), respectively. Deletions of the N-terminal Pex3-binding site disrupted the direct interactions of Pex19 with Pex3, but preserved interactions with a membrane peroxisomal targeting signal (mPTS)-containing PMP, Pex10. In contrast, deletion of the C-terminal mPTS-binding domain of Pex19 disrupted its interaction with Pex10 while leaving the Pex19-Pex3 interactions intact. However, Pex11 and Pex25 retained their interactions with both N- and C-terminal deletion mutants. NTC-CTC co-expression improved growth and reversed the larger-than-normal peroxisome size observed with the single deletions. Pex25 was critical for peroxisome formation with the CTC variants, and its overexpression enhanced their interactions with Pex3 and aided the growth of both NTC and CTC Pex19 variants. In conclusion, physical segregation of the Pex3- and PMP-binding domains of Pex19 has provided novel insights into the modular architecture of Pex19. We define the minimum region of Pex19 required for peroxisome biogenesis and a unique role for Pex25 in this process.
Keywords: membrane biogenesis; organelle; peroxisome; protein assembly; trafficking.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Subramani S. (1998) Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol. Rev. 78, 171–188 - PubMed
-
- Wanders R. J., and Waterham H. R. (2006) Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332 - PubMed
-
- Schrader M., and Fahimi H. D. (2006) Growth and division of peroxisomes. Int. Rev. Cytol. 255, 237–290 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

