Pseudomonas chlororaphis Produces Two Distinct R-Tailocins That Contribute to Bacterial Competition in Biofilms and on Roots
- PMID: 28526791
- PMCID: PMC5514687
- DOI: 10.1128/AEM.00706-17
Pseudomonas chlororaphis Produces Two Distinct R-Tailocins That Contribute to Bacterial Competition in Biofilms and on Roots
Abstract
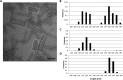
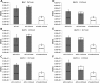
R-type tailocins are high-molecular-weight bacteriocins that resemble bacteriophage tails and are encoded within the genomes of many Pseudomonas species. In this study, analysis of the P. chlororaphis 30-84 R-tailocin gene cluster revealed that it contains the structural components to produce two R-tailocins of different ancestral origins. Two distinct R-tailocin populations differing in length were observed in UV-induced lysates of P. chlororaphis 30-84 via transmission electron microscopy. Mutants defective in the production of one or both R-tailocins demonstrated that the killing spectrum of each tailocin is limited to Pseudomonas species. The spectra of pseudomonads killed by the two R-tailocins differed, although a few Pseudomonas species were either killed by or insusceptible to both tailocins. Tailocin release was disrupted by deletion of the holin gene within the tailocin gene cluster, demonstrating that the lysis cassette is required for the release of both R-tailocins. The loss of functional tailocin production reduced the ability of P. chlororaphis 30-84 to compete with an R-tailocin-sensitive strain within biofilms and rhizosphere communities. Our study demonstrates that Pseudomonas species can produce more than one functional R-tailocin particle sharing the same lysis cassette but differing in their killing spectra. This study provides evidence for the role of R-tailocins as determinants of bacterial competition among plant-associated Pseudomonas in biofilms and the rhizosphere.IMPORTANCE Recent studies have identified R-tailocin gene clusters potentially encoding more than one R-tailocin within the genomes of plant-associated Pseudomonas but have not demonstrated that more than one particle is produced or the ecological significance of the production of multiple R-tailocins. This study demonstrates for the first time that Pseudomonas strains can produce two distinct R-tailocins with different killing spectra, both of which contribute to bacterial competition between rhizosphere-associated bacteria. These results provide new insight into the previously uncharacterized role of R-tailocin production by plant-associated Pseudomonas species in bacterial population dynamics within surface-attached biofilms and on roots.
Keywords: Pseudomonas; R-tailocin; bacterial competition; bacteriocins; microbial ecology; rhizosphere; rhizosphere-inhabiting microbes.
Copyright © 2017 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

