CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo
- PMID: 28527722
- PMCID: PMC5542791
- DOI: 10.1016/j.ymthe.2017.04.027
CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo
Abstract
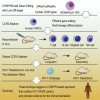
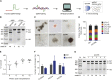
Transplantation of hematopoietic stem cells (HSCs) with a naturally occurring CCR5 mutation confers a loss of detectable HIV-1 in the patient, making ablation of the CCR5 gene in HSCs an ideal therapy for an HIV-1 cure. Although CCR5 disruption has been attempted in CD4+ T cells and hematopoietic stem/progenitor cells (HSPCs), efficient gene editing with high specificity and long-term therapeutic potential remains a major challenge for clinical translation. Here, we established a CRISPR/Cas9 gene editing system in human CD34+ HSPCs and achieved efficient CCR5 ablation evaluated in long-term reconstituted NOD/Prkdcscid/IL-2Rγnull mice. The CCR5 disruption efficiency in our system remained robust in secondary transplanted repopulating hematopoietic cells. More importantly, an HIV-1 resistance effect was observed as indicated by significant reduction of virus titration and enrichment of human CD4+ T cells. Hence, we successfully established a CRISPR/Cas9 mediated CCR5 ablating system in long-term HSCs, which confers HIV-1 resistance in vivo. Our study provides evidence for translating CCR5 gene-edited HSC transplantation for an HIV cure to the clinic.
Keywords: CCR5; CRISPR; HIV-1/AIDS; gene editing; gene therapy; hematopoietic stem cell.
Copyright © 2017 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures


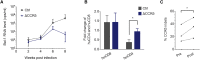
References
-
- Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R.E., Hill C.M. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. - PubMed
-
- Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. - PubMed
-
- Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R., MacDonald M.E., Stuhlmann H., Koup R.A., Landau N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. - PubMed
-
- Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. - PubMed
-
- Allers K., Hütter G., Hofmann J., Loddenkemper C., Rieger K., Thiel E., Schneider T. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

