Biochemical principles and inhibitors to interfere with viral capping pathways
- PMID: 28527860
- PMCID: PMC7185569
- DOI: 10.1016/j.coviro.2017.04.003
Biochemical principles and inhibitors to interfere with viral capping pathways
Abstract
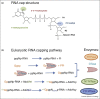
Messenger RNAs are decorated by a cap structure, which is essential for their translation into proteins. Many viruses have developed strategies in order to cap their mRNAs. The cap is either synthetized by a subset of viral or cellular enzymes, or stolen from capped cellular mRNAs by viral endonucleases ('cap-snatching'). Reverse genetic studies provide evidence that inhibition of viral enzymes belonging to the capping pathway leads to inhibition of virus replication. The replication defect results from reduced protein synthesis as well as from detection of incompletely capped RNAs by cellular innate immunity sensors. Thus, it is now admitted that capping enzymes are validated antiviral targets, as their inhibition will support an antiviral response in addition to the attenuation of viral mRNA translation. In this review, we describe the different viral enzymes involved in mRNA capping together with relevant inhibitors, and their biochemical features useful in inhibitor discovery.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures


References
-
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

