An Embryonic Stem Cell-Specific NuRD Complex Functions through Interaction with WDR5
- PMID: 28528697
- PMCID: PMC5470077
- DOI: 10.1016/j.stemcr.2017.04.020
An Embryonic Stem Cell-Specific NuRD Complex Functions through Interaction with WDR5
Abstract
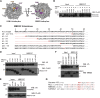
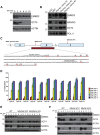
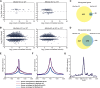
The Nucleosome Remodeling and Deacetylase (NuRD) complex is a chromatin regulatory complex that functions as a transcriptional co-repressor in metazoans. The NuRD subunit MBD3 is essential for targeting and assembly of a functional NuRD complex as well as embryonic stem cell (ESC) pluripotency. Three MBD3 isoforms (MBD3A, MBD3B, and MBD3C) are expressed in mouse. Here, we find that the MBD3C isoform contains a unique 50-amino-acid N-terminal region that is necessary for MBD3C to specifically interact with the histone H3 binding protein WDR5. Domain analyses of WDR5 reveal that the H3 binding pocket is required for interaction with MBD3C. We find that while Mbd3c knockout ESCs differentiate normally, MBD3C is redundant with the MBD3A and MBD3B isoforms in regulation of gene expression, with the unique MBD3C N terminus required for this redundancy. Together, our data characterize a unique NuRD complex variant that functions specifically in ESCs.
Keywords: Mbd3; NuRD; Wdr5; chromatin; differentiation.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

