Expansion and Purification Are Critical for the Therapeutic Application of Pluripotent Stem Cell-Derived Myogenic Progenitors
- PMID: 28528701
- PMCID: PMC5511038
- DOI: 10.1016/j.stemcr.2017.04.022
Expansion and Purification Are Critical for the Therapeutic Application of Pluripotent Stem Cell-Derived Myogenic Progenitors
Abstract
Recent reports have documented the differentiation of human pluripotent stem cells toward the skeletal myogenic lineage using transgene- and cell purification-free approaches. Although these protocols generate myocytes, they have not demonstrated scalability, safety, and in vivo engraftment, which are key aspects for their future clinical application. Here we recapitulate one prominent protocol, and show that it gives rise to a heterogeneous cell population containing myocytes and other cell types. Upon transplantation, the majority of human donor cells could not contribute to myofiber formation. As a proof-of-principle, we incorporated the inducible PAX7 lentiviral system into this protocol, which then enabled scalable expansion of a homogeneous population of skeletal myogenic progenitors capable of forming myofibers in vivo. Our findings demonstrate the methods for scalable expansion of PAX7+ myogenic progenitors and their purification are critical for practical application to cell replacement treatment of muscle degenerative diseases.
Keywords: MHC(+) myocytes; PAX7(+) myogenic progenitors; engraftment; morphogens; pluripotent stem cells; purification; skeletal muscle; stem cell therapy; transgene.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
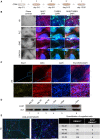
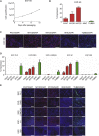
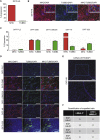
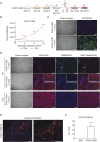
References
-
- Barberi T., Bradbury M., Dincer Z., Panagiotakos G., Socci N.D., Studer L. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat. Med. 2007;13:642–648. - PubMed
-
- Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., Bousson F., Zidouni Y., Mursch C., Moncuquet P. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotech. 2015;33:962–969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

