A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations
- PMID: 28528867
- PMCID: PMC5502825
- DOI: 10.1016/j.ccell.2017.04.013
A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations
Abstract
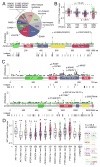
Molecular alterations involving the PI3K/AKT/mTOR pathway (including mutation, copy number, protein, or RNA) were examined across 11,219 human cancers representing 32 major types. Within specific mutated genes, frequency, mutation hotspot residues, in silico predictions, and functional assays were all informative in distinguishing the subset of genetic variants more likely to have functional relevance. Multiple oncogenic pathways including PI3K/AKT/mTOR converged on similar sets of downstream transcriptional targets. In addition to mutation, structural variations and partial copy losses involving PTEN and STK11 showed evidence for having functional relevance. A substantial fraction of cancers showed high mTOR pathway activity without an associated canonical genetic or genomic alteration, including cancers harboring IDH1 or VHL mutations, suggesting multiple mechanisms for pathway activation.
Keywords: PI3K/AKT/mTOR pathway; The Cancer Genome Atlas; integrative genomics analysis; pan-cancer analysis; proteomics; reverse-phase protein arrays.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures






References
-
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. - PubMed
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- P50 CA101942/CA/NCI NIH HHS/United States
- U24 CA210950/CA/NCI NIH HHS/United States
- P01 CA120964/CA/NCI NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- U01 CA168394/CA/NCI NIH HHS/United States
- U01 CA217842/CA/NCI NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- R01 CA175486/CA/NCI NIH HHS/United States
- U24 CA210949/CA/NCI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- R21 CA191687/CA/NCI NIH HHS/United States
- U24 CA209851/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

