Methodologies for studying the spliceosome's RNA dynamics with single-molecule FRET
- PMID: 28529063
- PMCID: PMC5546994
- DOI: 10.1016/j.ymeth.2017.05.011
Methodologies for studying the spliceosome's RNA dynamics with single-molecule FRET
Abstract
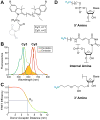
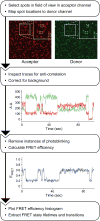
The spliceosome is an extraordinarily dynamic molecular machine in which significant changes in composition as well as protein and RNA conformation are required for carrying out pre-mRNA splicing. Single-molecule fluorescence resonance energy transfer (smFRET) can be used to elucidate these dynamics both in well-characterized model systems and in entire spliceosomes. These types of single-molecule data provide novel information about spliceosome components and can be used to identify sub-populations of molecules with unique behaviors. When smFRET is combined with single-molecule fluorescence colocalization, conformational dynamics can be further linked to the presence or absence of a given spliceosome component. Here, we provide a description of experimental considerations, approaches, and workflows for smFRET with an emphasis on applications for the splicing machinery.
Keywords: Dynamics; FRET; Fluorescence; RNA; Single-molecule; Spliceosome.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





References
-
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. - PubMed
-
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. - PubMed
-
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349(6253):1182–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

