Characterization of Cytosine Methylation and the DNA Methyltransferases of Toxoplasma gondii
- PMID: 28529454
- PMCID: PMC5436566
- DOI: 10.7150/ijbs.18644
Characterization of Cytosine Methylation and the DNA Methyltransferases of Toxoplasma gondii
Abstract
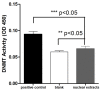
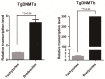
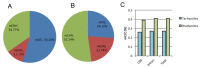
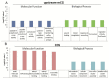
DNA methylation is a key epigenetic modification which confers phenotypic plasticity and adaptation. Cyst-forming strains of Toxoplasma gondii undergo tachyzoite to bradyzoite conversion after initial acute infection of a host, and the reverse conversion may occur in immune-suppressed hosts. The formation of m5C is catalyzed by DNA methyltransferase (DNMT). We identified two functional DNA methyltransferases, TgDNMTa and TgDNMTb, in T. gondii that may mediate DNA methylation. The recombinant proteins showed intrinsic methyltransferase activity; both have higher transcription levels in bradyzoites than that in tachyzoites. We performed genome-wide analysis of DNA methylation in tachyzoites and bradyzoites. The results showed more methylation sites in bradyzoites than that in tachyzoites. The most significantly enriched GO-terms of genes with DNA methylation were associated with basal cellular processes such as energy metabolism and parasite resistance to host immunity. Tachyzoite proliferation in parasitophorous vacuoles (PV) can be inhibited by the DNA methyltransferase inhibitor 5-azacytidine, a chemical analogue of the nucleotide cytosine that can inactivate DNA methyltransferases. These findings provide the first confirmation of DNA methylation in T. gondii.
Keywords: 5-azacytidine.; 5-cytosine methyltransferase; DNA methylation; Toxoplasma gondii; bradyzoite; tachyzoite.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




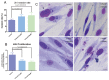
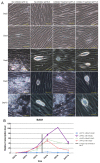
Similar articles
-
Toxoplasma gondii DNA methyltransferases regulate parasitic energy metabolism.Acta Trop. 2022 May;229:106329. doi: 10.1016/j.actatropica.2022.106329. Epub 2022 Feb 2. Acta Trop. 2022. PMID: 35122712
-
Characterization of protein arginine methyltransferase of TgPRMT5 in Toxoplasma gondii.Parasit Vectors. 2019 May 8;12(1):221. doi: 10.1186/s13071-019-3464-1. Parasit Vectors. 2019. PMID: 31068219 Free PMC article.
-
Toxoplasma gondii Cyclic AMP-Dependent Protein Kinase Subunit 3 Is Involved in the Switch from Tachyzoite to Bradyzoite Development.mBio. 2016 May 31;7(3):e00755-16. doi: 10.1128/mBio.00755-16. mBio. 2016. PMID: 27247232 Free PMC article.
-
The differential expression of multiple isoenzyme forms during stage conversion of Toxoplasma gondii: an adaptive developmental strategy.Int J Parasitol. 2001 Aug;31(10):1023-31. doi: 10.1016/s0020-7519(01)00193-x. Int J Parasitol. 2001. PMID: 11429165 Review.
-
The Epigenome, Cell Cycle, and Development in Toxoplasma.Annu Rev Microbiol. 2018 Sep 8;72:479-499. doi: 10.1146/annurev-micro-090817-062741. Epub 2018 Jun 22. Annu Rev Microbiol. 2018. PMID: 29932347 Review.
Cited by
-
The Absence of C-5 DNA Methylation in Leishmania donovani Allows DNA Enrichment from Complex Samples.Microorganisms. 2020 Aug 18;8(8):1252. doi: 10.3390/microorganisms8081252. Microorganisms. 2020. PMID: 32824654 Free PMC article.
-
"Reading" a new chapter in protozoan parasite transcriptional regulation.PLoS Pathog. 2021 Dec 2;17(12):e1010056. doi: 10.1371/journal.ppat.1010056. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34855919 Free PMC article. Review.
-
Two distinct SWI/SNF complexes direct chromatin-linked transcriptional programs in Toxoplasma.bioRxiv [Preprint]. 2025 Jul 16:2025.07.16.665172. doi: 10.1101/2025.07.16.665172. bioRxiv. 2025. PMID: 40791499 Free PMC article. Preprint.
-
Non‐m6A RNA modifications in haematological malignancies.Clin Transl Med. 2024 Jun;14(6):e1666. doi: 10.1002/ctm2.1666. Clin Transl Med. 2024. PMID: 38880983 Free PMC article. Review. Italian.
-
Cross-lineage 5-methylcytosine methylome profiling reveals methylated divergence among Toxoplasma gondii tachyzoites of the three major clonal lineages.Infect Dis Poverty. 2025 Aug 19;14(1):87. doi: 10.1186/s40249-025-01358-w. Infect Dis Poverty. 2025. PMID: 40830525 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

