Microdissection of the Ah01 chromosome in upland cotton and microcloning of resistance gene anologs from the single chromosome
- PMID: 28529470
- PMCID: PMC5437636
- DOI: 10.1186/s41065-017-0035-3
Microdissection of the Ah01 chromosome in upland cotton and microcloning of resistance gene anologs from the single chromosome
Abstract
Background: Chromosome microdissection is one of the most important techniques in molecular cytogenetic research. Cotton (Gossypium Linnaeus, 1753) is the main natural fiber crop in the world. The resistance gene analog (RGA) cloning after its single chromosome microdissection can greatly promote cotton genome research and breeding.
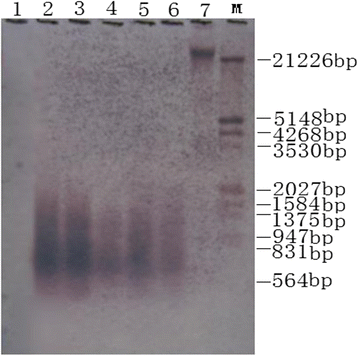
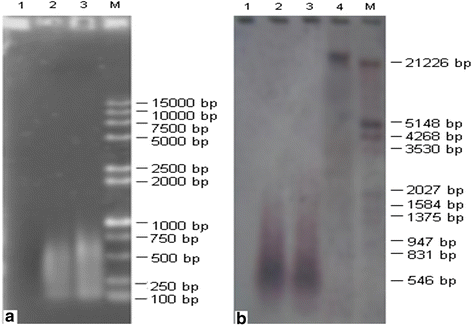
Results: Using the linker adaptor PCR (LA-PCR) with the primers of rice disease-resistance homologues, three nucleotide sequences PS016 (KU051681), PS054 (KU051682), and PS157 (KU051680) were obtained from the chromosome Ah01 of upland cotton (cv. TM-1). The Blast results showed that the three sequences are the nucleotide binding site-leucine rich repeat (NBS-LRR) type RGAs. Clustering results indicated that they are homologous to these published RGAs. Thus, the three RGAs can definitely be confirmed as NBS-LRR class of RGAs in upland cotton.
Conclusions: Using single chromosome microdissection technique, DNA libraries containing cotton RGAs were obtained. This technique can promote cotton gene cloning, marker development and even the improvement of cotton genome research and breeding.
Keywords: Chromosome microdissection; Microcloning; RGA; Upland cotton.
Figures





Similar articles
-
Resistance gene analogue markers are mapped to homeologous chromosomes in cultivated tetraploid cotton.Theor Appl Genet. 2005 Apr;110(6):1074-85. doi: 10.1007/s00122-005-1928-5. Epub 2005 Feb 22. Theor Appl Genet. 2005. PMID: 15726317
-
Comparative analysis of resistance gene analogues encoding NBS-LRR domains in cotton.J Sci Food Agric. 2016 Jan 30;96(2):530-8. doi: 10.1002/jsfa.7120. Epub 2015 Mar 3. J Sci Food Agric. 2016. PMID: 25640313
-
Cloning of resistance gene analogs located on the alien chromosome in an addition line of wheat-Thinopyrum intermedium.Theor Appl Genet. 2005 Sep;111(5):923-31. doi: 10.1007/s00122-005-0022-3. Epub 2005 Oct 18. Theor Appl Genet. 2005. PMID: 16044269
-
Disease Resistance Gene Analogs (RGAs) in Plants.Int J Mol Sci. 2015 Aug 14;16(8):19248-90. doi: 10.3390/ijms160819248. Int J Mol Sci. 2015. PMID: 26287177 Free PMC article. Review.
-
The Elusive Search for Reniform Nematode Resistance in Cotton.Phytopathology. 2018 May;108(5):532-541. doi: 10.1094/PHYTO-09-17-0320-RVW. Epub 2018 Feb 5. Phytopathology. 2018. PMID: 29116883 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

