Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases
- PMID: 28529637
- PMCID: PMC5436513
- DOI: 10.7150/thno.15625
Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases
Abstract
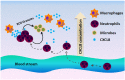
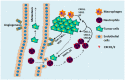
The chemokine receptors CXCR1/2 and their ligand CXCL8 are essential for the activation and trafficking of inflammatory mediators as well as tumor progression and metastasis. The CXCL8-CXCR1/2 signaling axis is involved in the pathogenesis of several diseases including chronic obstructive pulmonary diseases (COPD), asthma, cystic fibrosis and cancer. Interaction between CXCL8 secreted by select cancer cells and CXCR1/2 in the tumor microenvironment is critical for cancer progression and metastasis. The CXCL8-CXCR1/2 axis may play an important role in tumor progression and metastasis by regulating cancer stem cell (CSC) proliferation and self-renewal. During the past two decades, several small-molecule CXCR1/2 inhibitors, CXCL8 releasing inhibitors, and neutralizing antibodies against CXCL8 and CXCR1/2 have been reported. As single agents, such inhibitors are expected to be efficacious in various inflammatory diseases. Several preclinical studies suggest that combination of CXCR1/2 inhibitors along with other targeted therapies, chemotherapies, and immunotherapy may be effective in treating select cancers. Currently, several of these inhibitors are in advanced clinical trials for COPD, asthma, and metastatic breast cancer. In this review, we provide a comprehensive analysis of the role of the CXCL8-CXCR1/2 axis and select genes co-expressed in this pathway in disease progression. We also discuss the latest progress in developing small-molecule drugs targeting this pathway.
Keywords: CXCL8; CXCR1; CXCR2; antibody.; cancer; cancer stem cells; chronic obstructive pulmonary diseases; inhibitor; tumor microenvironment.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures











References
-
- Le Y, Zhou Y, Iribarren P, Wang J. Chemokines and chemokine receptors: their manifold roles in homeostasis and disease. Cell Mol Immunol. 2004;1:95–104. - PubMed
-
- Rollins BJ. Chemokines. Blood. 1997;90:909–28. - PubMed
-
- Hebert CA, Vitangcol RV, Baker JB. Scanning mutagenesis of interleukin-8 identifies a cluster of residues required for receptor binding. J Biol Chem. 1991;266:18989–94. - PubMed
-
- GPCRdb. http://gpcrdb.org/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

