Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations
- PMID: 28530384
- PMCID: PMC5832362
- DOI: 10.1021/acs.biochem.6b01192
Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations
Abstract
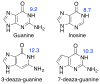
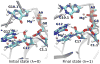
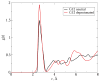
The hammerhead ribozyme is a well-studied nucleolytic ribozyme that catalyzes the self-cleavage of the RNA phosphodiester backbone. Despite experimental and theoretical efforts, key questions remain about details of the mechanism with regard to the activation of the nucleophile by the putative general base guanine (G12). Straightforward interpretation of the measured activity-pH data implies the pKa value of the N1 position in the G12 nucleobase is significantly shifted by the ribozyme environment. Recent crystallographic and biochemical work has identified pH-dependent divalent metal ion binding at the N7/O6 position of G12, leading to the hypothesis that this binding mode could induce a pKa shift of G12 toward neutrality. We present computational results that support this hypothesis and provide a model that unifies the interpretation of available structural and biochemical data and paints a detailed mechanistic picture of the general base step of the reaction. Experimentally testable predictions are made for mutational and rescue effects on G12, which will give further insights into the catalytic mechanism. These results contribute to our growing knowledge of the potential roles of divalent metal ions in RNA catalysis.
Figures




Similar articles
-
Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction.Biochemistry. 2015 Oct 20;54(41):6369-81. doi: 10.1021/acs.biochem.5b00824. Epub 2015 Oct 2. Biochemistry. 2015. PMID: 26398724 Free PMC article.
-
Existence of efficient divalent metal ion-catalyzed and inefficient divalent metal ion-independent channels in reactions catalyzed by a hammerhead ribozyme.Nucleic Acids Res. 2002 Jun 1;30(11):2374-82. doi: 10.1093/nar/30.11.2374. Nucleic Acids Res. 2002. PMID: 12034824 Free PMC article.
-
Molecular simulations of the pistol ribozyme: unifying the interpretation of experimental data and establishing functional links with the hammerhead ribozyme.RNA. 2019 Nov;25(11):1439-1456. doi: 10.1261/rna.071944.119. Epub 2019 Jul 30. RNA. 2019. PMID: 31363004 Free PMC article.
-
Folding and activity of the hammerhead ribozyme.Chembiochem. 2002 Aug 2;3(8):690-700. doi: 10.1002/1439-7633(20020802)3:8<690::AID-CBIC690>3.0.CO;2-C. Chembiochem. 2002. PMID: 12203967 Review.
-
Crystallographic structures of the hammerhead ribozyme: relationship to ribozyme folding and catalysis.Annu Rev Biophys Biomol Struct. 1998;27:475-502. doi: 10.1146/annurev.biophys.27.1.475. Annu Rev Biophys Biomol Struct. 1998. PMID: 9646875 Review.
Cited by
-
Hammerhead Ribozymes: Structural Insights, Catalytic Mechanisms, and Cutting-Edge Applications in Synthetic Biology.Int J Mol Sci. 2025 Jun 12;26(12):5624. doi: 10.3390/ijms26125624. Int J Mol Sci. 2025. PMID: 40565088 Free PMC article. Review.
-
Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes.Curr Opin Chem Biol. 2017 Dec;41:71-83. doi: 10.1016/j.cbpa.2017.09.017. Epub 2017 Nov 3. Curr Opin Chem Biol. 2017. PMID: 29107885 Free PMC article. Review.
-
Graph deep learning locates magnesium ions in RNA.QRB Discov. 2022;3:e20. doi: 10.1017/qrd.2022.17. Epub 2022 Oct 6. QRB Discov. 2022. PMID: 37292390 Free PMC article.
-
Catalytic Mechanism of Non-Target DNA Cleavage in CRISPR-Cas9 Revealed by Ab Initio Molecular Dynamics.ACS Catal. 2020 Nov 20;10(22):13596-13605. doi: 10.1021/acscatal.0c03566. Epub 2020 Nov 10. ACS Catal. 2020. PMID: 33520346 Free PMC article.
-
Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8-17 DNAzyme in solution.Nucleic Acids Res. 2019 Nov 4;47(19):10282-10295. doi: 10.1093/nar/gkz773. Nucleic Acids Res. 2019. PMID: 31511899 Free PMC article.
References
-
- Scott WG. RNA catalysis. Curr Opin Struct Biol. 1998;8:720–726. - PubMed
-
- Kuimelis RG, McLaughlin LW. Mechanisms of Ribozyme-Mediated RNA Cleavage. Chem Rev. 1998;98:1027–1044. - PubMed
-
- Zhou DM, Taira K. The Hydrolysis of RNA: From Theoretical Calculations to the Hammerhead Ribozyme-Mediated Cleavage of RNA. Chem Rev. 1998;98:991–1026. - PubMed
-
- Scott WG. Biophysical and biochemical investigations of RNA catalysis in the hammerhead ribozyme. Q Rev Biophys. 1999;32:241–294. - PubMed
-
- Takagi Y, Ikeda Y, Taira K. Ribozyme Mechanisms. Top Curr Chem. 2004;232:213–251.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

