Reevaluation of SNP heritability in complex human traits
- PMID: 28530675
- PMCID: PMC5493198
- DOI: 10.1038/ng.3865
Reevaluation of SNP heritability in complex human traits
Abstract
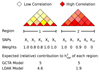
SNP heritability, the proportion of phenotypic variance explained by SNPs, has been reported for many hundreds of traits. Its estimation requires strong prior assumptions about the distribution of heritability across the genome, but current assumptions have not been thoroughly tested. By analyzing imputed data for a large number of human traits, we empirically derive a model that more accurately describes how heritability varies with minor allele frequency (MAF), linkage disequilibrium (LD) and genotype certainty. Across 19 traits, our improved model leads to estimates of common SNP heritability on average 43% (s.d. 3%) higher than those obtained from the widely used software GCTA and 25% (s.d. 2%) higher than those from the recently proposed extension GCTA-LDMS. Previously, DNase I hypersensitivity sites were reported to explain 79% of SNP heritability; using our improved heritability model, their estimated contribution is only 24%.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

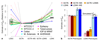

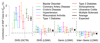
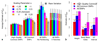
References
-
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. - PubMed
-
- Henderson C, Kempthorne O, Searle S, von Krosigk C. The estimation of environmental and genetic trends from records subject to culling. Biometrics. 1959;15:192–218.
-
- Falconer D, Mackay T. Introduction to Quantitative Genetics. 4th Edition. Longman; 1996.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

