BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency
- PMID: 28530713
- PMCID: PMC5593426
- DOI: 10.1038/ni.3753
BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency
Abstract
The transcriptional programs that guide lymphocyte differentiation depend on the precise expression and timing of transcription factors (TFs). The TF BACH2 is essential for T and B lymphocytes and is associated with an archetypal super-enhancer (SE). Single-nucleotide variants in the BACH2 locus are associated with several autoimmune diseases, but BACH2 mutations that cause Mendelian monogenic primary immunodeficiency have not previously been identified. Here we describe a syndrome of BACH2-related immunodeficiency and autoimmunity (BRIDA) that results from BACH2 haploinsufficiency. Affected subjects had lymphocyte-maturation defects that caused immunoglobulin deficiency and intestinal inflammation. The mutations disrupted protein stability by interfering with homodimerization or by causing aggregation. We observed analogous lymphocyte defects in Bach2-heterozygous mice. More generally, we observed that genes that cause monogenic haploinsufficient diseases were substantially enriched for TFs and SE architecture. These findings reveal a previously unrecognized feature of SE architecture in Mendelian diseases of immunity: heterozygous mutations in SE-regulated genes identified by whole-exome/genome sequencing may have greater significance than previously recognized.
Conflict of interest statement
Figures

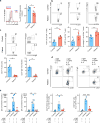

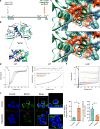

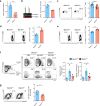
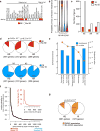
References
MeSH terms
Substances
Grants and funding
- MC_U120061454/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/000C0409/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P41 GM103311/GM/NIGMS NIH HHS/United States
- K22 HL125593/HL/NHLBI NIH HHS/United States
- 22597/CRUK_/Cancer Research UK/United Kingdom
- BB/N007794/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- Z99 HL999999/ImNIH/Intramural NIH HHS/United States
- BBS/E/B/000C0407/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

