Protein charge distribution in proteomes and its impact on translation
- PMID: 28531225
- PMCID: PMC5460897
- DOI: 10.1371/journal.pcbi.1005549
Protein charge distribution in proteomes and its impact on translation
Abstract
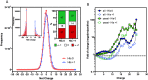
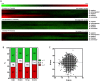
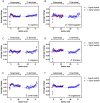
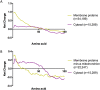
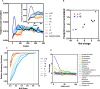
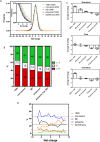
As proteins are synthesized, the nascent polypeptide must pass through a negatively charged exit tunnel. During this stage, positively charged stretches can interact with the ribosome walls and slow the translation. Therefore, charged polypeptides may be important factors that affect protein expression. To determine the frequency and distribution of positively and negatively charged stretches in different proteomes, the net charge was calculated for every 30 consecutive amino acid residues, which corresponds to the length of the ribosome exit tunnel. The following annotated and reviewed proteins in the UniProt database (Swiss-Prot) were analyzed: 551,705 proteins from different organisms and a total of 180 million protein segments. We observed that there were more negative than positive stretches and that super-charged positive sequences (i.e., net charges ≥ 14) were underrepresented in the proteomes. Overall, the proteins were more positively charged at their N-termini and C-termini, and this feature was present in most organisms and subcellular localizations. To investigate whether the N-terminal charges affect the elongation rates, previously published ribosomal profiling data obtained from S. cerevisiae, without translation-interfering drugs, were analyzed. We observed a nonlinear effect of the charge on the ribosome occupancy in which values ≥ +5 and ≤ -6 showed increased and reduced ribosome densities, respectively. These groups also showed different distributions across 80S monosomes and polysomes. Basic polypeptides are more common within short proteins that are translated by monosomes, whereas negative stretches are more abundant in polysome-translated proteins. These findings suggest that the nascent peptide charge impacts translation and can be one of the factors that regulate translation efficiency and protein expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

