Replication studies of carboxymethylated DNA lesions in human cells
- PMID: 28531304
- PMCID: PMC5499590
- DOI: 10.1093/nar/gkx442
Replication studies of carboxymethylated DNA lesions in human cells
Abstract
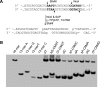
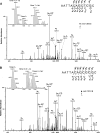
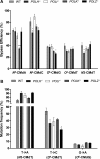
Metabolic activation of some N-nitroso compounds (NOCs), an important class of DNA damaging agents, can induce the carboxymethylation of nucleobases in DNA. Very little was previously known about how the carboxymethylated DNA lesions perturb DNA replication in human cells. Here, we investigated the effects of five carboxymethylated DNA lesions, i.e. O6-CMdG, N6-CMdA, N4-CMdC, N3-CMdT and O4-CMdT on the efficiency and fidelity of DNA replication in HEK293T human embryonic kidney cells. We found that, while neither N6-CMdA nor N4-CMdC blocked DNA replication or induced mutations, N3-CMdT, O4-CMdT and O6-CMdG moderately blocked DNA replication and induced substantial frequencies of T→A (81%), T→C (68%) and G→A (6.4%) mutations, respectively. In addition, our results revealed that CRISPR-Cas9-mediated depletion of Pol η resulted in significant drops in bypass efficiencies of N4-CMdC and N3-CMdT. Diminution in bypass efficiencies was also observed for N6-CMdA and O6-CMdG upon depletion of Pol κ, and for O6-CMdG upon removal of Pol ζ. Together, our study provided molecular-level insights into the impacts of the carboxymethylated DNA lesions on DNA replication in human cells, revealed the roles of individual translesion synthesis DNA polymerases in bypassing these lesions, and suggested the contributions of O6-CMdG, N3-CMdT and O4-CMdT to the mutations found in p53 gene of human gastrointestinal cancers.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
In vitro replication studies of carboxymethylated DNA lesions with Saccharomyces cerevisiae polymerase η.Biochemistry. 2011 Sep 6;50(35):7666-73. doi: 10.1021/bi2007417. Epub 2011 Aug 11. Biochemistry. 2011. PMID: 21809836 Free PMC article.
-
Translesion synthesis of O4-alkylthymidine lesions in human cells.Nucleic Acids Res. 2016 Nov 2;44(19):9256-9265. doi: 10.1093/nar/gkw662. Epub 2016 Jul 27. Nucleic Acids Res. 2016. PMID: 27466394 Free PMC article.
-
Transcriptional inhibition and mutagenesis induced by N-nitroso compound-derived carboxymethylated thymidine adducts in DNA.Nucleic Acids Res. 2015 Jan;43(2):1012-8. doi: 10.1093/nar/gku1391. Epub 2015 Jan 8. Nucleic Acids Res. 2015. PMID: 25572317 Free PMC article.
-
Mass Spectrometry-Based Quantitative Strategies for Assessing the Biological Consequences and Repair of DNA Adducts.Acc Chem Res. 2016 Feb 16;49(2):205-13. doi: 10.1021/acs.accounts.5b00437. Epub 2016 Jan 13. Acc Chem Res. 2016. PMID: 26758048 Free PMC article. Review.
-
DNA polymerase eta, a key protein in translesion synthesis in human cells.Subcell Biochem. 2010;50:189-209. doi: 10.1007/978-90-481-3471-7_10. Subcell Biochem. 2010. PMID: 20012583 Review.
Cited by
-
DNA replication studies of N-nitroso compound-induced O6-alkyl-2'-deoxyguanosine lesions in Escherichia coli.J Biol Chem. 2019 Mar 15;294(11):3899-3908. doi: 10.1074/jbc.RA118.007358. Epub 2019 Jan 17. J Biol Chem. 2019. PMID: 30655287 Free PMC article.
-
Chemical Analysis of DNA Damage.Anal Chem. 2018 Jan 2;90(1):556-576. doi: 10.1021/acs.analchem.7b04247. Epub 2017 Nov 7. Anal Chem. 2018. PMID: 29084424 Free PMC article. Review. No abstract available.
-
Biological Evaluation of DNA Biomarkers in a Chemically Defined and Site-Specific Manner.Toxics. 2019 Jun 25;7(2):36. doi: 10.3390/toxics7020036. Toxics. 2019. PMID: 31242562 Free PMC article. Review.
-
Formation of Carboxymethyl-Phosphotriester Adducts in DNA.Chem Res Toxicol. 2025 May 19;38(5):892-899. doi: 10.1021/acs.chemrestox.4c00547. Epub 2025 Apr 16. Chem Res Toxicol. 2025. PMID: 40235319
-
Detection and Discrimination of DNA Adducts Differing in Size, Regiochemistry, and Functional Group by Nanopore Sequencing.Chem Res Toxicol. 2020 Dec 21;33(12):2944-2952. doi: 10.1021/acs.chemrestox.0c00202. Epub 2020 Oct 1. Chem Res Toxicol. 2020. PMID: 32799528 Free PMC article.
References
-
- Louis P., Hold G.L., Flint H.J.. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014; 12:661–672. - PubMed
-
- Tricker A. N-nitroso compounds and man: sources of exposure, endogenous formation and occurrence in body fluids. Eur. J. Cancer Prev. 1997; 6:226–268. - PubMed
-
- Mirvish S.S. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 1995; 93:17–48. - PubMed
-
- Bingham S.A., Hughes R., Cross A.J.. Effect of white versus red meat on endogenous N-nitrosation in the human colon and further evidence of a dose response. J. Nutr. 2002; 132:3522S–3525S. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

