Structural Insights into the Eukaryotic Transcription Initiation Machinery
- PMID: 28532216
- PMCID: PMC6186020
- DOI: 10.1146/annurev-biophys-070816-033751
Structural Insights into the Eukaryotic Transcription Initiation Machinery
Abstract
Eukaryotic gene transcription requires the assembly at the promoter of a large preinitiation complex (PIC) that includes RNA polymerase II (Pol II) and the general transcription factors TFIID, TFIIA, TFIIB, TFIIF, TFIIE, and TFIIH. The size and complexity of Pol II, TFIID, and TFIIH have precluded their reconstitution from heterologous systems, and purification relies on scarce endogenous sources. Together with their conformational flexibility and the transient nature of their interactions, these limitations had precluded structural characterization of the PIC. In the last few years, however, progress in cryo-electron microscopy (cryo-EM) has made possible the visualization, at increasingly better resolution, of large PIC assemblies in different functional states. These structures can now be interpreted in near-atomic detail and provide an exciting structural framework for past and future functional studies, giving us unique mechanistic insight into the complex process of transcription initiation.
Keywords: PIC; TFIID; TFIIE; TFIIF; TFIIH; cryo-EM; general transcription factors.
Figures


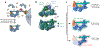


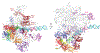

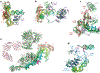


References
-
- Akoulitchev S, Mäkelä TP, Weinberg RA, Reinberg D. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557–60 - PubMed
-
- Albright SR, Tjian R. 2000. TAPs revisited: More data reveal new twists and confirm old ideas. Gene 242:1–13 - PubMed
-
- Andel F III, Ladurner AG, Inouye C, Tjian R, Nogales E. 1999. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286:2153–56 - PubMed
-
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, et al. 2013. The architecture of human general transcription factor TFIID core complex. Nature 493:699–702 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

