Robust estimation of the expected survival probabilities from high-dimensional Cox models with biomarker-by-treatment interactions in randomized clinical trials
- PMID: 28532387
- PMCID: PMC5441049
- DOI: 10.1186/s12874-017-0354-0
Robust estimation of the expected survival probabilities from high-dimensional Cox models with biomarker-by-treatment interactions in randomized clinical trials
Abstract
Background: Thanks to the advances in genomics and targeted treatments, more and more prediction models based on biomarkers are being developed to predict potential benefit from treatments in a randomized clinical trial. Despite the methodological framework for the development and validation of prediction models in a high-dimensional setting is getting more and more established, no clear guidance exists yet on how to estimate expected survival probabilities in a penalized model with biomarker-by-treatment interactions.
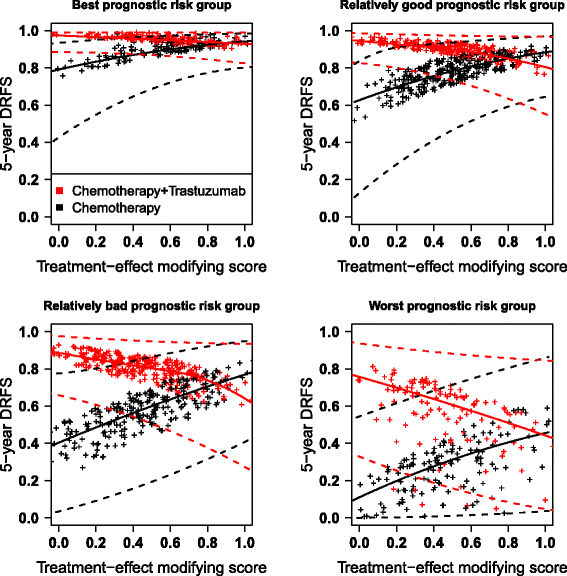
Methods: Based on a parsimonious biomarker selection in a penalized high-dimensional Cox model (lasso or adaptive lasso), we propose a unified framework to: estimate internally the predictive accuracy metrics of the developed model (using double cross-validation); estimate the individual survival probabilities at a given timepoint; construct confidence intervals thereof (analytical or bootstrap); and visualize them graphically (pointwise or smoothed with spline). We compared these strategies through a simulation study covering scenarios with or without biomarker effects. We applied the strategies to a large randomized phase III clinical trial that evaluated the effect of adding trastuzumab to chemotherapy in 1574 early breast cancer patients, for which the expression of 462 genes was measured.
Results: In our simulations, penalized regression models using the adaptive lasso estimated the survival probability of new patients with low bias and standard error; bootstrapped confidence intervals had empirical coverage probability close to the nominal level across very different scenarios. The double cross-validation performed on the training data set closely mimicked the predictive accuracy of the selected models in external validation data. We also propose a useful visual representation of the expected survival probabilities using splines. In the breast cancer trial, the adaptive lasso penalty selected a prediction model with 4 clinical covariates, the main effects of 98 biomarkers and 24 biomarker-by-treatment interactions, but there was high variability of the expected survival probabilities, with very large confidence intervals.
Conclusion: Based on our simulations, we propose a unified framework for: developing a prediction model with biomarker-by-treatment interactions in a high-dimensional setting and validating it in absence of external data; accurately estimating the expected survival probability of future patients with associated confidence intervals; and graphically visualizing the developed prediction model. All the methods are implemented in the R package biospear, publicly available on the CRAN.
Keywords: Confidence intervals; Cox model; High-dimensional data; Penalized regression; Precision medicine; Prediction model; Prognostic biomarkers; Survival estimation; Treatment-effect modifiers.
Figures



References
-
- Perez EA, Thompson EA, Ballman KV, Anderson SK, Asmann YW, Kalari KR, et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group n9831 Adjuvant Trastuzumab Trial. J Clin Oncol. 2015;33:701–708. doi: 10.1200/JCO.2014.57.6298. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

