A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing
- PMID: 28532419
- PMCID: PMC5440902
- DOI: 10.1186/s12864-017-3757-8
A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing
Abstract
Background: Despite the economic importance of sugarcane in sugar and bioenergy production, there is not yet a reference genome available. Most of the sugarcane transcriptomic studies have been based on Saccharum officinarum gene indices (SoGI), expressed sequence tags (ESTs) and de novo assembled transcript contigs from short-reads; hence knowledge of the sugarcane transcriptome is limited in relation to transcript length and number of transcript isoforms.
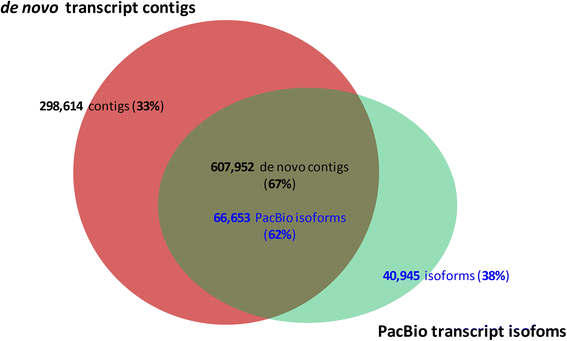
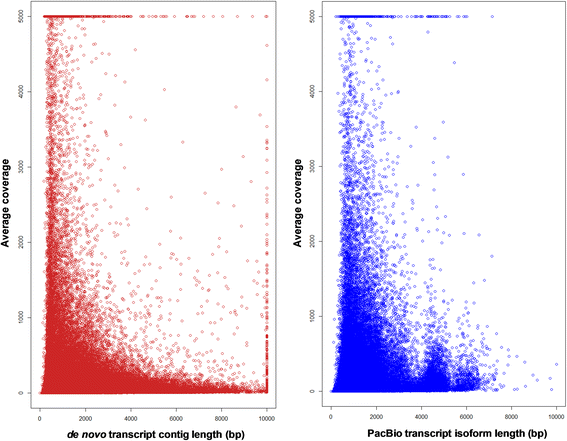
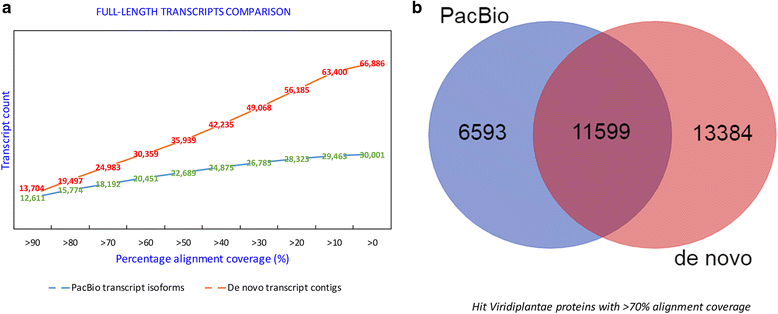
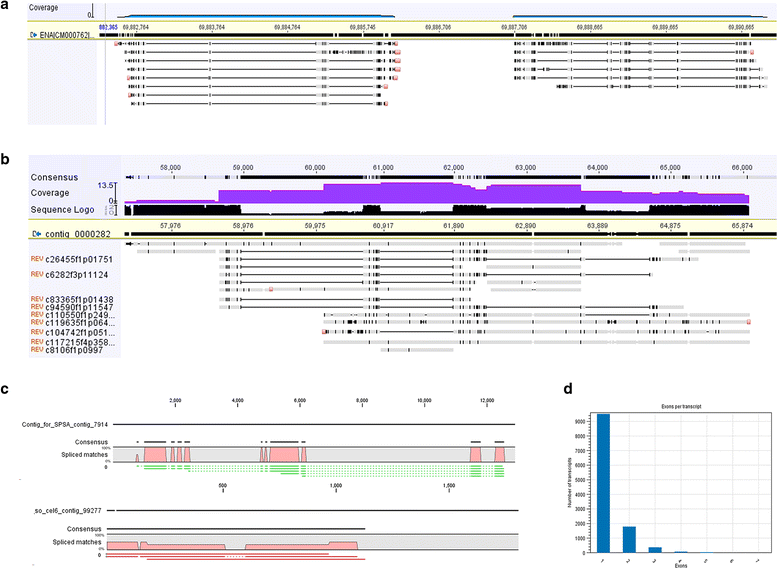
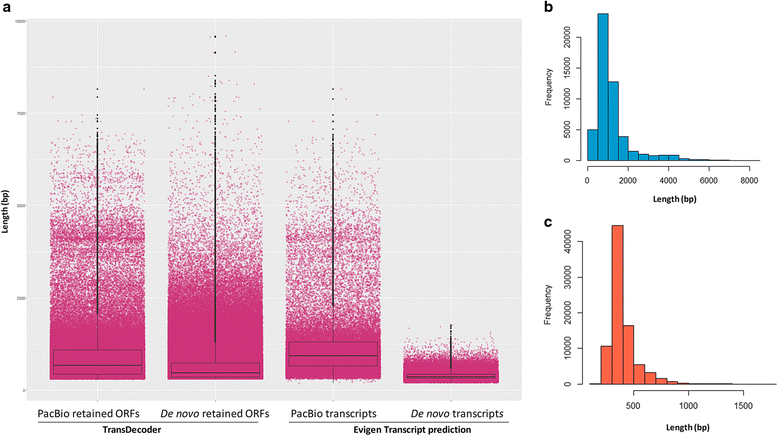
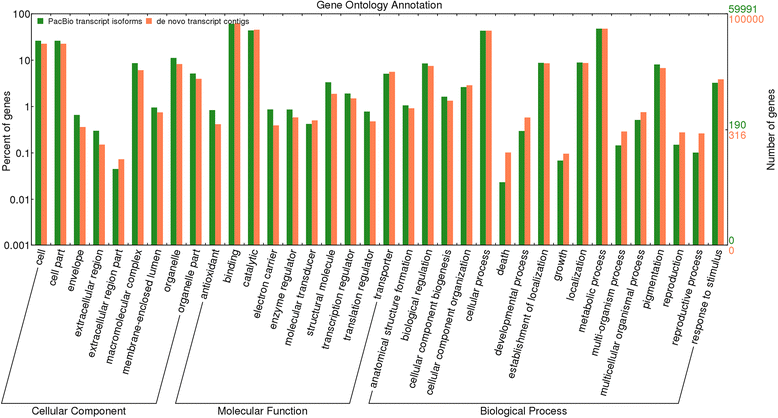
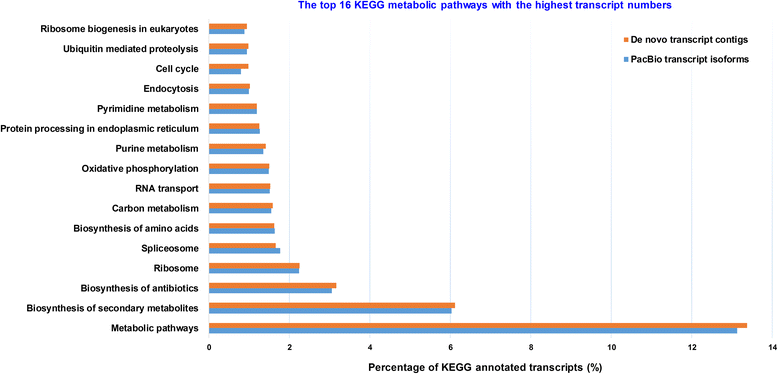
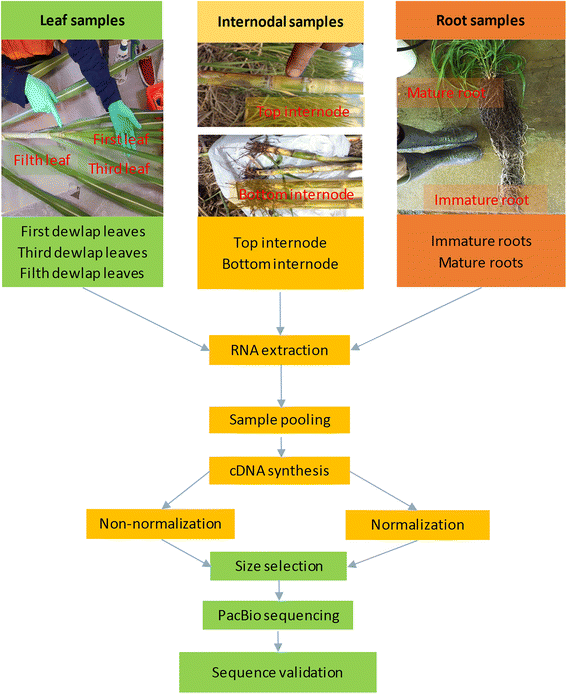
Results: The sugarcane transcriptome was sequenced using PacBio isoform sequencing (Iso-Seq) of a pooled RNA sample derived from leaf, internode and root tissues, of different developmental stages, from 22 varieties, to explore the potential for capturing full-length transcript isoforms. A total of 107,598 unique transcript isoforms were obtained, representing about 71% of the total number of predicted sugarcane genes. The majority of this dataset (92%) matched the plant protein database, while just over 2% was novel transcripts, and over 2% was putative long non-coding RNAs. About 56% and 23% of total sequences were annotated against the gene ontology and KEGG pathway databases, respectively. Comparison with de novo contigs from Illumina RNA-Sequencing (RNA-Seq) of the internode samples from the same experiment and public databases showed that the Iso-Seq method recovered more full-length transcript isoforms, had a higher N50 and average length of largest 1,000 proteins; whereas a greater representation of the gene content and RNA diversity was captured in RNA-Seq. Only 62% of PacBio transcript isoforms matched 67% of de novo contigs, while the non-matched proportions were attributed to the inclusion of leaf/root tissues and the normalization in PacBio, and the representation of more gene content and RNA classes in the de novo assembly, respectively. About 69% of PacBio transcript isoforms and 41% of de novo contigs aligned with the sorghum genome, indicating the high conservation of orthologs in the genic regions of the two genomes.
Conclusions: The transcriptome dataset should contribute to improved sugarcane gene models and sugarcane protein predictions; and will serve as a reference database for analysis of transcript expression in sugarcane.
Keywords: De novo assembly; Hybrid assembly; Isoform sequencing; Polyploid transcriptome; SUGIT database; Sugarcane; Transcriptome assembly.
Figures

Similar articles
-
Unveiling the transcriptomic complexity of Miscanthus sinensis using a combination of PacBio long read- and Illumina short read sequencing platforms.BMC Genomics. 2021 Sep 22;22(1):690. doi: 10.1186/s12864-021-07971-x. BMC Genomics. 2021. PMID: 34551715 Free PMC article.
-
Uncovering full-length transcript isoforms of sugarcane cultivar Khon Kaen 3 using single-molecule long-read sequencing.PeerJ. 2018 Oct 30;6:e5818. doi: 10.7717/peerj.5818. eCollection 2018. PeerJ. 2018. PMID: 30397543 Free PMC article.
-
A de novo Full-Length mRNA Transcriptome Generated From Hybrid-Corrected PacBio Long-Reads Improves the Transcript Annotation and Identifies Thousands of Novel Splice Variants in Atlantic Salmon.Front Genet. 2021 Apr 27;12:656334. doi: 10.3389/fgene.2021.656334. eCollection 2021. Front Genet. 2021. PMID: 33986770 Free PMC article.
-
PacBio Sequencing and Its Applications.Genomics Proteomics Bioinformatics. 2015 Oct;13(5):278-89. doi: 10.1016/j.gpb.2015.08.002. Epub 2015 Nov 2. Genomics Proteomics Bioinformatics. 2015. PMID: 26542840 Free PMC article. Review.
-
The Challenge of Analyzing the Sugarcane Genome.Front Plant Sci. 2018 May 14;9:616. doi: 10.3389/fpls.2018.00616. eCollection 2018. Front Plant Sci. 2018. PMID: 29868072 Free PMC article. Review.
Cited by
-
Full-length SMRT transcriptome sequencing and microsatellite characterization in Paulownia catalpifolia.Sci Rep. 2021 Apr 22;11(1):8734. doi: 10.1038/s41598-021-87538-8. Sci Rep. 2021. PMID: 33888729 Free PMC article.
-
Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content.Sci Rep. 2018 Aug 2;8(1):11612. doi: 10.1038/s41598-018-30033-4. Sci Rep. 2018. PMID: 30072760 Free PMC article.
-
Full Transcriptome Analysis of Callus Suspension Culture System of Bletilla striata.Front Genet. 2020 Oct 15;11:995. doi: 10.3389/fgene.2020.00995. eCollection 2020. Front Genet. 2020. PMID: 33193583 Free PMC article.
-
A hybrid correcting method considering heterozygous variations by a comprehensive probabilistic model.BMC Genomics. 2020 Nov 18;21(Suppl 10):753. doi: 10.1186/s12864-020-07008-9. BMC Genomics. 2020. PMID: 33208104 Free PMC article.
-
Investigation of RNA Editing Sites within Bound Regions of RNA-Binding Proteins.High Throughput. 2019 Nov 29;8(4):19. doi: 10.3390/ht8040019. High Throughput. 2019. PMID: 31795425 Free PMC article.
References
-
- Hotta C, Lembke C, Domingues D, Ochoa E, Cruz GQ, Melotto-Passarin D, Marconi T, Santos M, Mollinari M, Margarido GA, et al. The biotechnology roadmap for sugarcane improvement. Trop Plant Biol. 2010;3(2):75–87. doi: 10.1007/s12042-010-9050-5. - DOI
-
- Vettore AL, da Silva FR, Kemper EL, Souza GM, da Silva AM, Ferro MI, Henrique-Silva F, Giglioti EA, Lemos MV, Coutinho LL, et al. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003;13(12):2725–2735. doi: 10.1101/gr.1532103. - DOI - PMC - PubMed
-
- Souza GM, Berges H, Bocs S, Casu R, D’Hont A, Ferreira JE, Henry R, Ming R, Potier B, Sluys M-A, et al. The sugarcane genome challenge: strategies for sequencing a highly complex genome. Trop Plant Biol. 2011;4(3–4):145–156. doi: 10.1007/s12042-011-9079-0. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

