Research Techniques Made Simple: High-Throughput Sequencing of the T-Cell Receptor
- PMID: 28532760
- PMCID: PMC5912324
- DOI: 10.1016/j.jid.2017.04.001
Research Techniques Made Simple: High-Throughput Sequencing of the T-Cell Receptor
Abstract
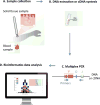
High-throughput sequencing (HTS) of the T-cell receptor (TCR) is a rapidly advancing technique that allows sensitive and accurate identification and quantification of every distinct T-cell clone present within any biological sample. The relative frequency of each individual clone within the full T-cell repertoire can also be studied. HTS is essential to expand our knowledge on the diversity of the TCR repertoire in homeostasis or under pathologic conditions, as well as to understand the kinetics of antigen-specific T-cell responses that lead to protective immunity (i.e., vaccination) or immune-related disorders (i.e., autoimmunity and cancer). HTS can be tailored for personalized medicine, having the potential to monitor individual responses to therapeutic interventions and show prognostic and diagnostic biomarkers. In this article, we briefly review the methodology, advances, and limitations of HTS of the TCR and describe emerging applications of this technique in the field of investigative dermatology. We highlight studying the pathogenesis of T cells in allergic dermatitis and the application of HTS of the TCR in diagnosing, detecting recurrence early, and monitoring responses to therapy in cutaneous T-cell lymphoma.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
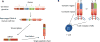


References
-
- Chitgopeker P, Sahni D. T-Cell Receptor Gene Rearrangement Detection in Suspected Cases of Cutaneous T-Cell Lymphoma. J Invest Dermatol. 2014;134:e19. - PubMed
-
- Chung WH, Pan RY, Chu MT, Chin SW, Huang YL, Wang WC, et al. Oxypurinol-Specific T Cells Possess Preferential TCR Clonotypes and Express Granulysin in Allopurinol-Induced Severe Cutaneous Adverse Reactions. J Invest Dermatol. 2015 Sep;135(9):2237–48. - PubMed
-
- Howie B, Sherwood AM, Berkebile AD, Berka J, Emerson RO, Williamson DW, et al. High-throughput pairing of T cell receptor α and β sequences. Sci Transl Med. 2015 Aug 19;7(301):301ra131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

