Enhancer-derived RNA: A Primer
- PMID: 28533025
- PMCID: PMC5487531
- DOI: 10.1016/j.gpb.2016.12.006
Enhancer-derived RNA: A Primer
Abstract
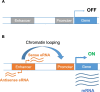
Enhancer-derived RNAs (eRNAs) are a group of RNAs transcribed by RNA polymerase II from the domain of transcription enhancers, a major type of cis-regulatory elements in the genome. The correlation between eRNA production and enhancer activity has stimulated studies on the potential role of eRNAs in transcriptional regulation. Additionally, eRNA has also served as a marker for global identification of enhancers. Here I review the brief history and fascinating properties of eRNAs.
Keywords: Enhancer; Gene regulation; Next-generation sequencing; Non-coding RNA; eRNA.
Copyright © 2017 The Author. Production and hosting by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Non-coding RNA derived from the region adjacent to the human HO-1 E2 enhancer selectively regulates HO-1 gene induction by modulating Pol II binding.Nucleic Acids Res. 2014 Dec 16;42(22):13599-614. doi: 10.1093/nar/gku1169. Epub 2014 Nov 17. Nucleic Acids Res. 2014. PMID: 25404134 Free PMC article.
-
Enhancer RNAs step forward: new insights into enhancer function.Development. 2022 Aug 15;149(16):dev200398. doi: 10.1242/dev.200398. Epub 2022 Aug 30. Development. 2022. PMID: 36039999 Free PMC article.
-
Transcriptional control by enhancers and enhancer RNAs.Transcription. 2019 Aug-Oct;10(4-5):171-186. doi: 10.1080/21541264.2019.1695492. Epub 2019 Dec 2. Transcription. 2019. PMID: 31791217 Free PMC article.
-
Insights into Enhancer RNAs: Biogenesis and Emerging Role in Brain Diseases.Neuroscientist. 2023 Apr;29(2):166-176. doi: 10.1177/10738584211046889. Epub 2021 Oct 6. Neuroscientist. 2023. PMID: 34612730 Review.
-
Enhancer RNA: biogenesis, function, and regulation.Essays Biochem. 2020 Dec 7;64(6):883-894. doi: 10.1042/EBC20200014. Essays Biochem. 2020. PMID: 33034351 Review.
Cited by
-
A statistical framework for predicting critical regions of p53-dependent enhancers.Brief Bioinform. 2021 May 20;22(3):bbaa053. doi: 10.1093/bib/bbaa053. Brief Bioinform. 2021. PMID: 32392580 Free PMC article.
-
Cis-Regulatory Elements in Mammals.Int J Mol Sci. 2023 Dec 26;25(1):343. doi: 10.3390/ijms25010343. Int J Mol Sci. 2023. PMID: 38203513 Free PMC article. Review.
-
Master control: transcriptional regulation of mammalian Myod.J Muscle Res Cell Motil. 2019 Jun;40(2):211-226. doi: 10.1007/s10974-019-09538-6. Epub 2019 Jul 12. J Muscle Res Cell Motil. 2019. PMID: 31301002 Free PMC article. Review.
-
Enhancer occlusion transcripts regulate the activity of human enhancer domains via transcriptional interference: a computational perspective.Nucleic Acids Res. 2020 Apr 17;48(7):3435-3454. doi: 10.1093/nar/gkaa026. Nucleic Acids Res. 2020. PMID: 32133533 Free PMC article.
-
Characterization of GECPAR, a noncoding RNA that regulates the transcriptional program of diffuse large B-cell lymphoma.Haematologica. 2022 May 1;107(5):1131-1143. doi: 10.3324/haematol.2020.267096. Haematologica. 2022. PMID: 34162177 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

